
Simple-effect analysis and post-hoc multiple comparison.
Source:R/bruceR-stats_3_manova.R
EMMEANS.RdPerform (1) simple-effect (and simple-simple-effect) analyses, including both simple main effects and simple interaction effects, and (2) post-hoc multiple comparisons (e.g., pairwise, sequential, polynomial), with p values adjusted for factors with >= 3 levels. This function is based on and extends emmeans::joint_tests(), emmeans::emmeans(), and emmeans::contrast(). You only need to specify the model object, to-be-tested effect(s), and moderator(s). Almost all results you need will be displayed together, including effect sizes (partial \(\eta^2\) and Cohen's d) and their confidence intervals (CIs). 90% CIs for partial \(\eta^2\) and 95% CIs for Cohen's d are reported.
Usage
EMMEANS(
model,
effect = NULL,
by = NULL,
contrast = "pairwise",
reverse = TRUE,
p.adjust = "bonferroni",
sd.pooled = NULL,
model.type = "multivariate",
digits = 3,
file = NULL
)Arguments
- model
The model object returned by
MANOVA().- effect
Effect(s) you want to test. If set to a character string (e.g.,
"A"), it reports the results of omnibus test or simple main effect. If set to a character vector (e.g.,c("A", "B")), it also reports the results of simple interaction effect.- by
Moderator variable(s). Defaults to
NULL.- contrast
Contrast method for multiple comparisons. Defaults to
"pairwise".Options:
"pairwise","revpairwise","seq","consec","poly","eff". For details, see emmeans::contrast-methods.- reverse
The order of levels to be contrasted. Defaults to
TRUE(higher level vs. lower level).- p.adjust
Adjustment method of p values for multiple comparisons. Defaults to
"bonferroni". For polynomial contrasts, defaults to"none".Options:
"none","fdr","hochberg","hommel","holm","tukey","mvt","dunnettx","sidak","scheffe","bonferroni". For details, seestats::p.adjust()andemmeans::summary.emmGrid().- sd.pooled
By default, it uses
sqrt(MSE)(root mean square error, RMSE) as the pooled SD to compute Cohen's d. Users may specify this argument as the SD of a reference group, or useeffectsize::sd_pooled()to obtain a pooled SD. For an issue about the computation method of Cohen's d, see the Disclaimer section.- model.type
"multivariate"returns the results of pairwise comparisons identical to SPSS, which uses thelm(rather thanaov) object of themodelforemmeans::joint_tests()andemmeans::emmeans()."univariate"requires also specifyingaov.include=TRUEinMANOVA(), which is not recommended by theafexpackage, seeafex::aov_ez().- digits
Number of decimal places of output. Defaults to
3.- file
File name of MS Word (
".doc").
Value
The same model object as returned by MANOVA() (for recursive use), with a list of tables: sim (simple effects), emm (estimated marginal means), con (contrasts). Each EMMEANS(...) appends one list to the returned object.
Disclaimer
By default, the root mean square error (RMSE) is used to compute the pooled SD for Cohen's d. Specifically, it uses:
the square root of mean square error (MSE) for between-subjects designs;
the square root of mean variance of all paired differences of the residuals of repeated measures for within-subjects and mixed designs.
Disclaimer: There is substantial disagreement on the appropriate pooled SD to use in computing the effect size. For alternative methods, see emmeans::eff_size() and effectsize::t_to_d(). Please do not take the default output as the only right results and users are completely responsible for specifying sd.pooled.
Interaction Plot
You can save the returned object and use the emmeans::emmip() function to create an interaction plot (based on the fitted model and a formula). See examples below for the usage. emmeans::emmip() returns a ggplot object, which can be modified and saved with ggplot2 syntax.
Statistical Details
Some may confuse the statistical terms "simple effects", "post-hoc tests", and "multiple comparisons". Such a confusion is not uncommon. Here I explain what these terms actually refer to.
1. Simple Effect
When we speak of "simple effect", we are referring to ...
simple main effect
simple interaction effect (only for designs with 3 or more factors)
simple simple effect (only for designs with 3 or more factors)
When the interaction effect in ANOVA is significant, we should then perform a "simple-effect analysis". In regression, we call this "simple-slope analysis". They are identical in statistical principles.
In a two-factors design, we only test "simple main effect". That is, at different levels of a factor "B", the main effects of "A" would be different. However, in a three-factors (or more) design, we may also test "simple interaction effect" and "simple simple effect". That is, at different combinations of levels of factors "B" and "C", the main effects of "A" would be different.
To note, simple effects per se never require p-value adjustment, because what we test in simple-effect analyses are still "omnibus F-tests".
2. Post-Hoc Test
The term "post-hoc" means that the tests are performed after ANOVA. Given this, some may (wrongly) regard simple-effect analyses also as a kind of post-hoc tests. However, these two terms should be distinguished. In many situations, "post-hoc tests" only refer to "post-hoc comparisons" using t-tests and some p-value adjustment techniques. We need post-hoc comparisons only when there are factors with 3 or more levels.
Post-hoc tests are totally independent of whether there is a significant interaction effect. It only deals with factors with multiple levels. In most cases, we use pairwise comparisons to do post-hoc tests. See the next part for details.
3. Multiple Comparison
As mentioned above, multiple comparisons are indeed post-hoc tests but have no relationship with simple-effect analyses. Post-hoc multiple comparisons are independent of interaction effects and simple effects. Furthermore, if a simple main effect contains 3 or more levels, we also need to do multiple comparisons within the simple-effect analysis. In this situation, we also need p-value adjustment with methods such as Bonferroni, Tukey's HSD (honest significant difference), FDR (false discovery rate), and so forth.
Options for multiple comparison:
"pairwise": Pairwise comparisons (defaults to "higher level - lower level")"seq"or"consec": Consecutive (sequential) comparisons"poly": Polynomial contrasts (linear, quadratic, cubic, quartic, ...)"eff": Effect contrasts (vs. the grand mean)
Examples
#### Between-Subjects Design ####
# \donttest{
between.1
#> A SCORE
#> 1 1 3
#> 2 1 6
#> 3 1 4
#> 4 1 3
#> 5 1 5
#> 6 1 7
#> 7 1 5
#> 8 1 2
#> 9 2 4
#> 10 2 6
#> 11 2 4
#> 12 2 2
#> 13 2 4
#> 14 2 5
#> 15 2 3
#> 16 2 3
#> 17 3 8
#> 18 3 9
#> 19 3 8
#> 20 3 7
#> 21 3 5
#> 22 3 6
#> 23 3 7
#> 24 3 6
#> 25 4 9
#> 26 4 8
#> 27 4 8
#> 28 4 7
#> 29 4 12
#> 30 4 13
#> 31 4 12
#> 32 4 11
MANOVA(between.1, dv="SCORE", between="A") %>%
EMMEANS("A")
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────
#> "A" Mean S.D. n
#> ─────────────────────
#> A1 4.375 (1.685) 8
#> A2 3.875 (1.246) 8
#> A3 7.000 (1.309) 8
#> A4 10.000 (2.268) 8
#> ─────────────────────
#> Total sample size: N = 32
#>
#> ANOVA Table:
#> Dependent variable(s): SCORE
#> Between-subjects factor(s): A
#> Within-subjects factor(s): –
#> Covariate(s): –
#> ─────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ─────────────────────────────────────────────────────────────────
#> A 63.375 2.813 3 28 22.533 <.001 *** .707 [.526, .798] .707
#> ─────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ───────────────────────────────────────
#> Levene’s F df1 df2 p
#> ───────────────────────────────────────
#> DV: SCORE 3.235 3 28 .037 *
#> ───────────────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────
#> Effect df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────
#> A 3 28 22.533 <.001 *** .707 [.526, .798]
#> ────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────
#> Mean: "A" 190.125 3 63.375 22.533 <.001 ***
#> Residuals 78.750 28 2.813
#> ─────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────
#> "A" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────
#> A1 4.375 [3.160, 5.590] (0.593)
#> A2 3.875 [2.660, 5.090] (0.593)
#> A3 7.000 [5.785, 8.215] (0.593)
#> A4 10.000 [8.785, 11.215] (0.593)
#> ───────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────
#> Contrast Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────
#> A2 - A1 -0.500 (0.839) 28 -0.596 1.000 -0.298 [-1.718, 1.121]
#> A3 - A1 2.625 (0.839) 28 3.130 .024 * 1.565 [ 0.146, 2.985]
#> A3 - A2 3.125 (0.839) 28 3.727 .005 ** 1.863 [ 0.444, 3.283]
#> A4 - A1 5.625 (0.839) 28 6.708 <.001 *** 3.354 [ 1.935, 4.774]
#> A4 - A2 6.125 (0.839) 28 7.304 <.001 *** 3.652 [ 2.233, 5.072]
#> A4 - A3 3.000 (0.839) 28 3.578 .008 ** 1.789 [ 0.369, 3.208]
#> ──────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.677
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
MANOVA(between.1, dv="SCORE", between="A") %>%
EMMEANS("A", p.adjust="tukey")
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────
#> "A" Mean S.D. n
#> ─────────────────────
#> A1 4.375 (1.685) 8
#> A2 3.875 (1.246) 8
#> A3 7.000 (1.309) 8
#> A4 10.000 (2.268) 8
#> ─────────────────────
#> Total sample size: N = 32
#>
#> ANOVA Table:
#> Dependent variable(s): SCORE
#> Between-subjects factor(s): A
#> Within-subjects factor(s): –
#> Covariate(s): –
#> ─────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ─────────────────────────────────────────────────────────────────
#> A 63.375 2.813 3 28 22.533 <.001 *** .707 [.526, .798] .707
#> ─────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ───────────────────────────────────────
#> Levene’s F df1 df2 p
#> ───────────────────────────────────────
#> DV: SCORE 3.235 3 28 .037 *
#> ───────────────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────
#> Effect df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────
#> A 3 28 22.533 <.001 *** .707 [.526, .798]
#> ────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────
#> Mean: "A" 190.125 3 63.375 22.533 <.001 ***
#> Residuals 78.750 28 2.813
#> ─────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────
#> "A" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────
#> A1 4.375 [3.160, 5.590] (0.593)
#> A2 3.875 [2.660, 5.090] (0.593)
#> A3 7.000 [5.785, 8.215] (0.593)
#> A4 10.000 [8.785, 11.215] (0.593)
#> ───────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────
#> Contrast Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────
#> A2 - A1 -0.500 (0.839) 28 -0.596 .932 -0.298 [-1.663, 1.067]
#> A3 - A1 2.625 (0.839) 28 3.130 .020 * 1.565 [ 0.200, 2.930]
#> A3 - A2 3.125 (0.839) 28 3.727 .005 ** 1.863 [ 0.498, 3.229]
#> A4 - A1 5.625 (0.839) 28 6.708 <.001 *** 3.354 [ 1.989, 4.719]
#> A4 - A2 6.125 (0.839) 28 7.304 <.001 *** 3.652 [ 2.287, 5.017]
#> A4 - A3 3.000 (0.839) 28 3.578 .007 ** 1.789 [ 0.424, 3.154]
#> ──────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.677
#> P-value adjustment: Tukey method for comparing a family of 4 estimates.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
MANOVA(between.1, dv="SCORE", between="A") %>%
EMMEANS("A", contrast="seq")
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────
#> "A" Mean S.D. n
#> ─────────────────────
#> A1 4.375 (1.685) 8
#> A2 3.875 (1.246) 8
#> A3 7.000 (1.309) 8
#> A4 10.000 (2.268) 8
#> ─────────────────────
#> Total sample size: N = 32
#>
#> ANOVA Table:
#> Dependent variable(s): SCORE
#> Between-subjects factor(s): A
#> Within-subjects factor(s): –
#> Covariate(s): –
#> ─────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ─────────────────────────────────────────────────────────────────
#> A 63.375 2.813 3 28 22.533 <.001 *** .707 [.526, .798] .707
#> ─────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ───────────────────────────────────────
#> Levene’s F df1 df2 p
#> ───────────────────────────────────────
#> DV: SCORE 3.235 3 28 .037 *
#> ───────────────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────
#> Effect df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────
#> A 3 28 22.533 <.001 *** .707 [.526, .798]
#> ────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────
#> Mean: "A" 190.125 3 63.375 22.533 <.001 ***
#> Residuals 78.750 28 2.813
#> ─────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────
#> "A" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────
#> A1 4.375 [3.160, 5.590] (0.593)
#> A2 3.875 [2.660, 5.090] (0.593)
#> A3 7.000 [5.785, 8.215] (0.593)
#> A4 10.000 [8.785, 11.215] (0.593)
#> ───────────────────────────────────
#>
#> Consecutive (Sequential) Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────
#> Contrast Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────
#> A2 - A1 -0.500 (0.839) 28 -0.596 1.000 -0.298 [-1.571, 0.975]
#> A3 - A2 3.125 (0.839) 28 3.727 .003 ** 1.863 [ 0.590, 3.137]
#> A4 - A3 3.000 (0.839) 28 3.578 .004 ** 1.789 [ 0.516, 3.062]
#> ──────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.677
#> P-value adjustment: Bonferroni method for 3 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
MANOVA(between.1, dv="SCORE", between="A") %>%
EMMEANS("A", contrast="poly")
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────
#> "A" Mean S.D. n
#> ─────────────────────
#> A1 4.375 (1.685) 8
#> A2 3.875 (1.246) 8
#> A3 7.000 (1.309) 8
#> A4 10.000 (2.268) 8
#> ─────────────────────
#> Total sample size: N = 32
#>
#> ANOVA Table:
#> Dependent variable(s): SCORE
#> Between-subjects factor(s): A
#> Within-subjects factor(s): –
#> Covariate(s): –
#> ─────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ─────────────────────────────────────────────────────────────────
#> A 63.375 2.813 3 28 22.533 <.001 *** .707 [.526, .798] .707
#> ─────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ───────────────────────────────────────
#> Levene’s F df1 df2 p
#> ───────────────────────────────────────
#> DV: SCORE 3.235 3 28 .037 *
#> ───────────────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────
#> Effect df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────
#> A 3 28 22.533 <.001 *** .707 [.526, .798]
#> ────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────
#> Mean: "A" 190.125 3 63.375 22.533 <.001 ***
#> Residuals 78.750 28 2.813
#> ─────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────
#> "A" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────
#> A1 4.375 [3.160, 5.590] (0.593)
#> A2 3.875 [2.660, 5.090] (0.593)
#> A3 7.000 [5.785, 8.215] (0.593)
#> A4 10.000 [8.785, 11.215] (0.593)
#> ───────────────────────────────────
#>
#> Polynomial Contrasts of "A":
#> ───────────────────────────────────────────────
#> Contrast Estimate S.E. df t p
#> ───────────────────────────────────────────────
#> linear 20.000 (2.652) 28 7.542 <.001 ***
#> quadratic 3.500 (1.186) 28 2.951 .006 **
#> cubic -3.750 (2.652) 28 -1.414 .168
#> ───────────────────────────────────────────────
#>
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
between.2
#> A B SCORE
#> 1 1 1 3
#> 2 1 1 6
#> 3 1 1 4
#> 4 1 1 3
#> 5 1 2 4
#> 6 1 2 6
#> 7 1 2 4
#> 8 1 2 2
#> 9 1 3 5
#> 10 1 3 7
#> 11 1 3 5
#> 12 1 3 2
#> 13 2 1 4
#> 14 2 1 5
#> 15 2 1 3
#> 16 2 1 3
#> 17 2 2 8
#> 18 2 2 9
#> 19 2 2 8
#> 20 2 2 7
#> 21 2 3 12
#> 22 2 3 13
#> 23 2 3 12
#> 24 2 3 11
MANOVA(between.2, dv="SCORE", between=c("A", "B")) %>%
EMMEANS("A", by="B") %>%
EMMEANS("B", by="A")
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────
#> "A" "B" Mean S.D. n
#> ─────────────────────────
#> A1 B1 4.000 (1.414) 4
#> A1 B2 4.000 (1.633) 4
#> A1 B3 4.750 (2.062) 4
#> A2 B1 3.750 (0.957) 4
#> A2 B2 8.000 (0.816) 4
#> A2 B3 12.000 (0.816) 4
#> ─────────────────────────
#> Total sample size: N = 24
#>
#> ANOVA Table:
#> Dependent variable(s): SCORE
#> Between-subjects factor(s): A, B
#> Within-subjects factor(s): –
#> Covariate(s): –
#> ─────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ─────────────────────────────────────────────────────────────────────
#> A 80.667 1.861 1 18 43.343 <.001 *** .707 [.482, .817] .707
#> B 40.542 1.861 2 18 21.784 <.001 *** .708 [.470, .815] .708
#> A * B 28.292 1.861 2 18 15.201 <.001 *** .628 [.347, .763] .628
#> ─────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ───────────────────────────────────────
#> Levene’s F df1 df2 p
#> ───────────────────────────────────────
#> DV: SCORE 0.605 5 18 .697
#> ───────────────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A B1 1 18 0.067 .798 .004 [.000, .137]
#> A B2 1 18 17.194 <.001 *** .489 [.198, .674]
#> A B3 1 18 56.485 <.001 *** .758 [.564, .849]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────
#> B1: "A" 0.125 1 0.125 0.067 .798
#> B2: "A" 32.000 1 32.000 17.194 <.001 ***
#> B3: "A" 105.125 1 105.125 56.485 <.001 ***
#> Residuals 33.500 18 1.861
#> ─────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 2.567, 5.433] (0.682)
#> A2 B1 3.750 [ 2.317, 5.183] (0.682)
#> A1 B2 4.000 [ 2.567, 5.433] (0.682)
#> A2 B2 8.000 [ 6.567, 9.433] (0.682)
#> A1 B3 4.750 [ 3.317, 6.183] (0.682)
#> A2 B3 12.000 [10.567, 13.433] (0.682)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 -0.250 (0.965) 18 -0.259 .798 -0.183 [-1.669, 1.302]
#> A2 - A1 B2 4.000 (0.965) 18 4.147 <.001 *** 2.932 [ 1.446, 4.418]
#> A2 - A1 B3 7.250 (0.965) 18 7.516 <.001 *** 5.314 [ 3.829, 6.800]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.364
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "B") ------
#>
#> Joint Tests of "B":
#> ────────────────────────────────────────────────────────
#> Effect "A" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> B A1 2 18 0.403 .674 .043 [.000, .205]
#> B A2 2 18 36.582 <.001 *** .803 [.631, .876]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "B":
#> ─────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────
#> A1: "B" 1.500 2 0.750 0.403 .674
#> A2: "B" 136.167 2 68.083 36.582 <.001 ***
#> Residuals 33.500 18 1.861
#> ─────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "B":
#> ────────────────────────────────────────
#> "B" "A" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> B1 A1 4.000 [ 2.567, 5.433] (0.682)
#> B2 A1 4.000 [ 2.567, 5.433] (0.682)
#> B3 A1 4.750 [ 3.317, 6.183] (0.682)
#> B1 A2 3.750 [ 2.317, 5.183] (0.682)
#> B2 A2 8.000 [ 6.567, 9.433] (0.682)
#> B3 A2 12.000 [10.567, 13.433] (0.682)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "B":
#> ─────────────────────────────────────────────────────────────────────────
#> Contrast "A" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ─────────────────────────────────────────────────────────────────────────
#> B2 - B1 A1 0.000 (0.965) 18 0.000 1.000 0.000 [-1.866, 1.866]
#> B3 - B1 A1 0.750 (0.965) 18 0.777 1.000 0.550 [-1.316, 2.416]
#> B3 - B2 A1 0.750 (0.965) 18 0.777 1.000 0.550 [-1.316, 2.416]
#> B2 - B1 A2 4.250 (0.965) 18 4.406 .001 ** 3.115 [ 1.249, 4.981]
#> B3 - B1 A2 8.250 (0.965) 18 8.552 <.001 *** 6.047 [ 4.181, 7.914]
#> B3 - B2 A2 4.000 (0.965) 18 4.147 .002 ** 2.932 [ 1.066, 4.798]
#> ─────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.364
#> P-value adjustment: Bonferroni method for 3 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
## How to create an interaction plot using `emmeans::emmip()`?
## See help page: ?emmeans::emmip()
m = MANOVA(between.2, dv="SCORE", between=c("A", "B"))
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────
#> "A" "B" Mean S.D. n
#> ─────────────────────────
#> A1 B1 4.000 (1.414) 4
#> A1 B2 4.000 (1.633) 4
#> A1 B3 4.750 (2.062) 4
#> A2 B1 3.750 (0.957) 4
#> A2 B2 8.000 (0.816) 4
#> A2 B3 12.000 (0.816) 4
#> ─────────────────────────
#> Total sample size: N = 24
#>
#> ANOVA Table:
#> Dependent variable(s): SCORE
#> Between-subjects factor(s): A, B
#> Within-subjects factor(s): –
#> Covariate(s): –
#> ─────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ─────────────────────────────────────────────────────────────────────
#> A 80.667 1.861 1 18 43.343 <.001 *** .707 [.482, .817] .707
#> B 40.542 1.861 2 18 21.784 <.001 *** .708 [.470, .815] .708
#> A * B 28.292 1.861 2 18 15.201 <.001 *** .628 [.347, .763] .628
#> ─────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ───────────────────────────────────────
#> Levene’s F df1 df2 p
#> ───────────────────────────────────────
#> DV: SCORE 0.605 5 18 .697
#> ───────────────────────────────────────
#>
emmip(m, ~ A | B, CIs=TRUE)
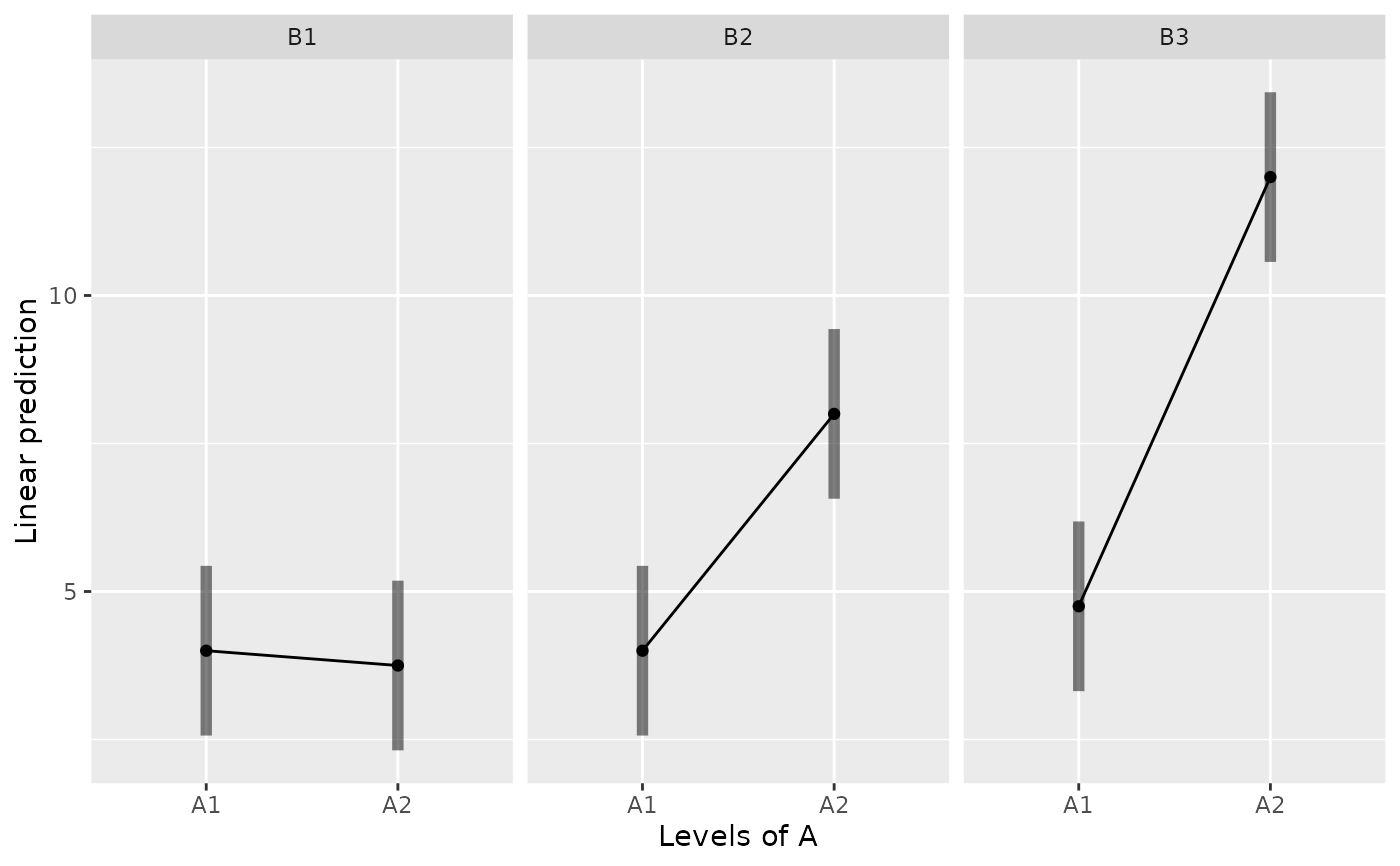 emmip(m, ~ B | A, CIs=TRUE)
emmip(m, ~ B | A, CIs=TRUE)
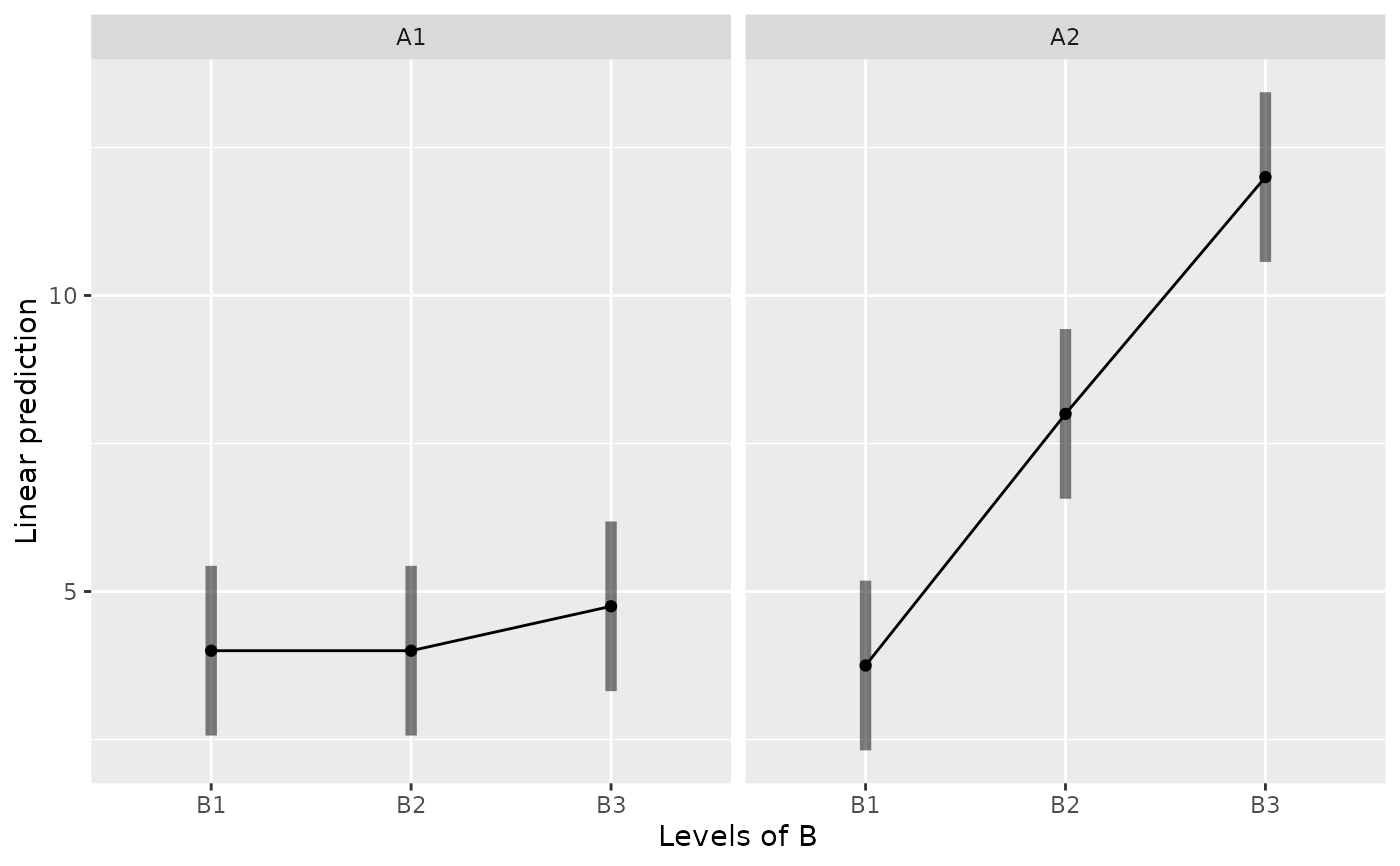 emmip(m, B ~ A, CIs=TRUE)
emmip(m, B ~ A, CIs=TRUE)
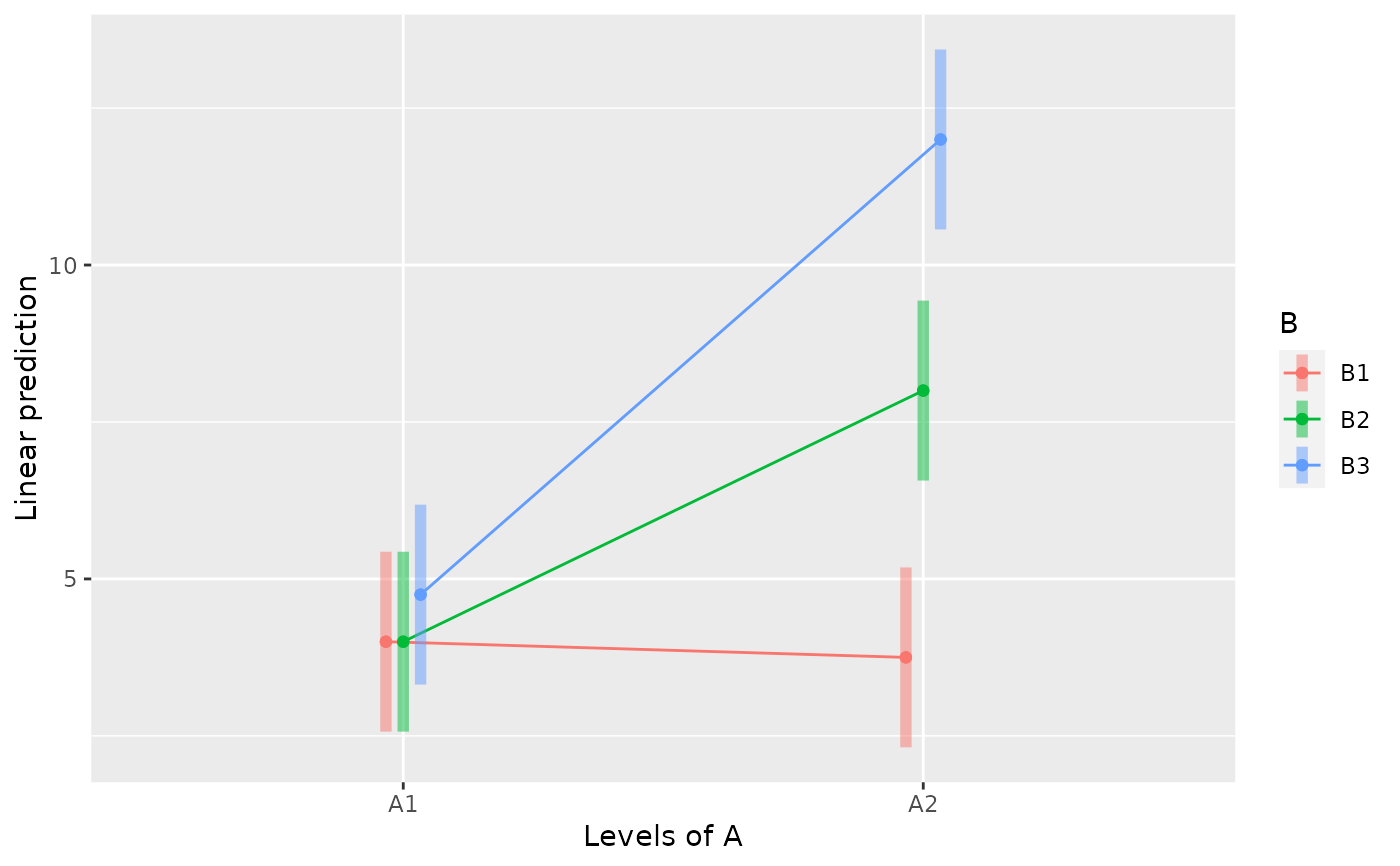 emmip(m, A ~ B, CIs=TRUE)
emmip(m, A ~ B, CIs=TRUE)
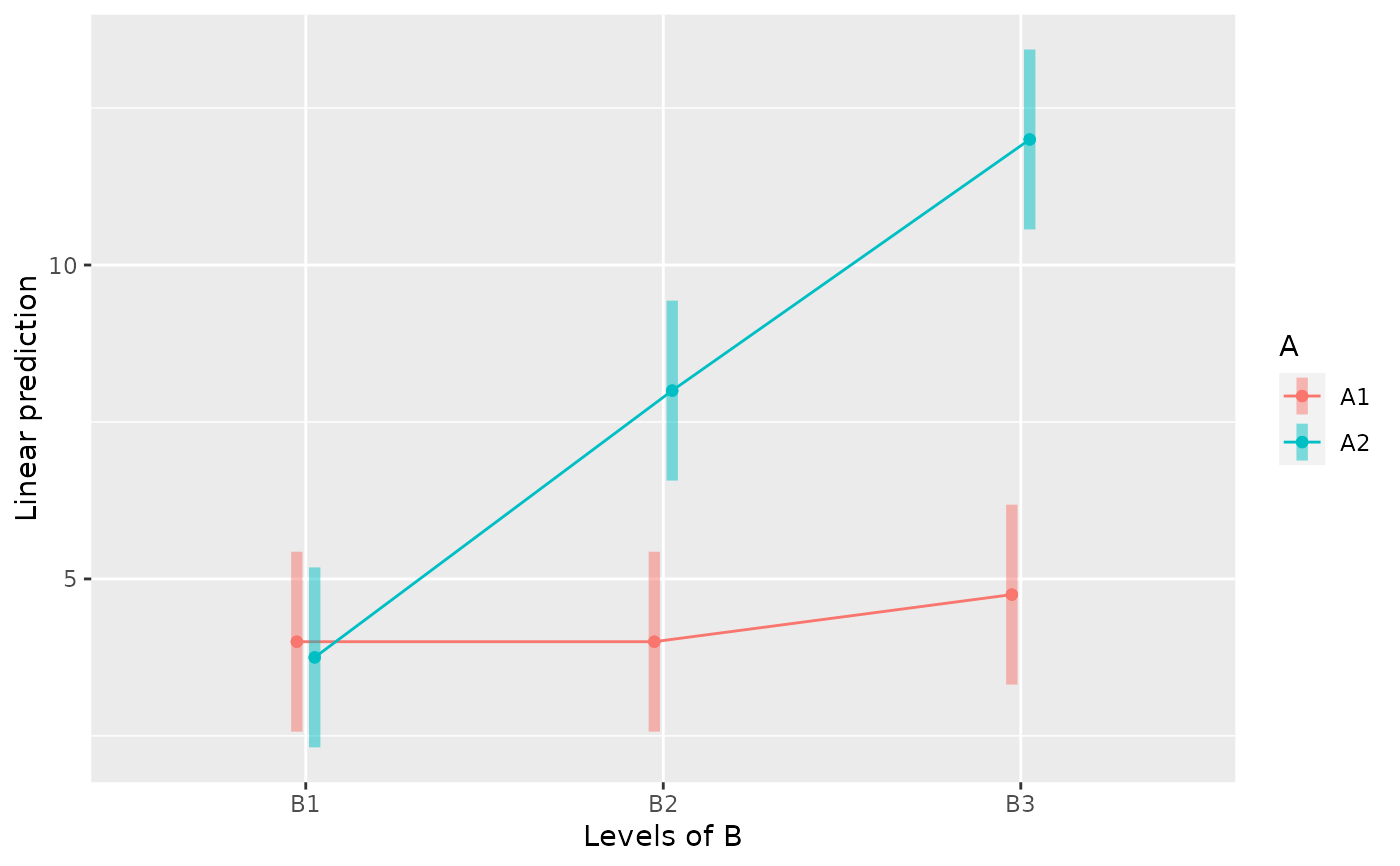 between.3
#> A B C SCORE
#> 1 1 1 1 3
#> 2 1 1 1 6
#> 3 1 1 1 4
#> 4 1 1 1 3
#> 5 1 1 2 5
#> 6 1 1 2 7
#> 7 1 1 2 5
#> 8 1 1 2 2
#> 9 1 2 1 4
#> 10 1 2 1 6
#> 11 1 2 1 4
#> 12 1 2 1 2
#> 13 1 2 2 4
#> 14 1 2 2 5
#> 15 1 2 2 3
#> 16 1 2 2 3
#> 17 2 1 1 8
#> 18 2 1 1 9
#> 19 2 1 1 8
#> 20 2 1 1 7
#> 21 2 1 2 5
#> 22 2 1 2 6
#> 23 2 1 2 7
#> 24 2 1 2 6
#> 25 2 2 1 9
#> 26 2 2 1 8
#> 27 2 2 1 8
#> 28 2 2 1 7
#> 29 2 2 2 12
#> 30 2 2 2 13
#> 31 2 2 2 12
#> 32 2 2 2 11
MANOVA(between.3, dv="SCORE", between=c("A", "B", "C")) %>%
EMMEANS("A", by="B") %>%
EMMEANS(c("A", "B"), by="C") %>%
EMMEANS("A", by=c("B", "C"))
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────────
#> "A" "B" "C" Mean S.D. n
#> ─────────────────────────────
#> A1 B1 C1 4.000 (1.414) 4
#> A1 B1 C2 4.750 (2.062) 4
#> A1 B2 C1 4.000 (1.633) 4
#> A1 B2 C2 3.750 (0.957) 4
#> A2 B1 C1 8.000 (0.816) 4
#> A2 B1 C2 6.000 (0.816) 4
#> A2 B2 C1 8.000 (0.816) 4
#> A2 B2 C2 12.000 (0.816) 4
#> ─────────────────────────────
#> Total sample size: N = 32
#>
#> ANOVA Table:
#> Dependent variable(s): SCORE
#> Between-subjects factor(s): A, B, C
#> Within-subjects factor(s): –
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 153.125 1.563 1 24 98.000 <.001 *** .803 [.670, .870] .803
#> B 12.500 1.563 1 24 8.000 .009 ** .250 [.042, .466] .250
#> C 3.125 1.563 1 24 2.000 .170 .077 [.000, .283] .077
#> A * B 24.500 1.563 1 24 15.680 <.001 *** .395 [.147, .585] .395
#> A * C 1.125 1.563 1 24 0.720 .405 .029 [.000, .206] .029
#> B * C 12.500 1.563 1 24 8.000 .009 ** .250 [.042, .466] .250
#> A * B * C 24.500 1.563 1 24 15.680 <.001 *** .395 [.147, .585] .395
#> ──────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ───────────────────────────────────────
#> Levene’s F df1 df2 p
#> ───────────────────────────────────────
#> DV: SCORE 0.668 7 24 .697
#> ───────────────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A B1 1 24 17.640 <.001 *** .424 [.173, .607]
#> A B2 1 24 96.040 <.001 *** .800 [.665, .868]
#> C B1 1 24 1.000 .327 .040 [.000, .226]
#> C B2 1 24 9.000 .006 ** .273 [.055, .486]
#> A * C B1 1 24 4.840 .038 * .168 [.006, .388]
#> A * C B2 1 24 11.560 .002 ** .325 [.090, .530]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────
#> B1: "A" 27.563 1 27.563 17.640 <.001 ***
#> B2: "A" 150.062 1 150.062 96.040 <.001 ***
#> Residuals 37.500 24 1.563
#> ─────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────
#> A1 B1 4.375 [3.463, 5.287] (0.442)
#> A2 B1 7.000 [6.088, 7.912] (0.442)
#> A1 B2 3.875 [2.963, 4.787] (0.442)
#> A2 B2 10.000 [9.088, 10.912] (0.442)
#> ───────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ─────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ─────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 2.625 (0.625) 24 4.200 <.001 *** 2.100 [1.068, 3.132]
#> A2 - A1 B2 6.125 (0.625) 24 9.800 <.001 *** 4.900 [3.868, 5.932]
#> ─────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.250
#> Results are averaged over the levels of: C
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A" & "B") ------
#>
#> Joint Tests of "A" & "B":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A C1 1 24 40.960 <.001 *** .631 [.414, .754]
#> A C2 1 24 57.760 <.001 *** .706 [.521, .806]
#> B C1 1 24 0.000 1.000 .000 [.000, .000]
#> B C2 1 24 16.000 <.001 *** .400 [.151, .589]
#> A * B C1 1 24 0.000 1.000 .000 [.000, .000]
#> A * B C2 1 24 31.360 <.001 *** .566 [.331, .710]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A" & "B":
#> ─────────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────────
#> C1: "A" & "B" 0.000 1 0.000 0.000 1.000
#> C2: "A" & "B" 49.000 1 49.000 31.360 <.001 ***
#> Residuals 37.500 24 1.563
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A" & "B":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 2.710, 5.290] (0.625)
#> A2 B1 8.000 [ 6.710, 9.290] (0.625)
#> A1 B2 4.000 [ 2.710, 5.290] (0.625)
#> A2 B2 8.000 [ 6.710, 9.290] (0.625)
#> A1 B1 4.750 [ 3.460, 6.040] (0.625)
#> A2 B1 6.000 [ 4.710, 7.290] (0.625)
#> A1 B2 3.750 [ 2.460, 5.040] (0.625)
#> A2 B2 12.000 [10.710, 13.290] (0.625)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A" & "B":
#> ───────────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ───────────────────────────────────────────────────────────────────────────────
#> A2 B1 - A1 B1 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.167, 5.233]
#> A1 B2 - A1 B1 C1 0.000 (0.884) 24 0.000 1.000 0.000 [-2.033, 2.033]
#> A1 B2 - A2 B1 C1 -4.000 (0.884) 24 -4.525 <.001 *** -3.200 [-5.233, -1.167]
#> A2 B2 - A1 B1 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.167, 5.233]
#> A2 B2 - A2 B1 C1 -0.000 (0.884) 24 -0.000 1.000 -0.000 [-2.033, 2.033]
#> A2 B2 - A1 B2 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.167, 5.233]
#> A2 B1 - A1 B1 C2 1.250 (0.884) 24 1.414 1.000 1.000 [-1.033, 3.033]
#> A1 B2 - A1 B1 C2 -1.000 (0.884) 24 -1.131 1.000 -0.800 [-2.833, 1.233]
#> A1 B2 - A2 B1 C2 -2.250 (0.884) 24 -2.546 .107 -1.800 [-3.833, 0.233]
#> A2 B2 - A1 B1 C2 7.250 (0.884) 24 8.202 <.001 *** 5.800 [ 3.767, 7.833]
#> A2 B2 - A2 B1 C2 6.000 (0.884) 24 6.788 <.001 *** 4.800 [ 2.767, 6.833]
#> A2 B2 - A1 B2 C2 8.250 (0.884) 24 9.334 <.001 *** 6.600 [ 4.567, 8.633]
#> ───────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.250
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────────
#> Effect "B" "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────────
#> A B1 C1 1 24 20.480 <.001 *** .460 [.210, .634]
#> A B2 C1 1 24 20.480 <.001 *** .460 [.210, .634]
#> A B1 C2 1 24 2.000 .170 .077 [.000, .283]
#> A B2 C2 1 24 87.120 <.001 *** .784 [.639, .858]
#> ────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A":
#> ────────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ────────────────────────────────────────────────────────────
#> B1 & C1: "A" 32.000 1 32.000 20.480 <.001 ***
#> B2 & C1: "A" 32.000 1 32.000 20.480 <.001 ***
#> B1 & C2: "A" 3.125 1 3.125 2.000 .170
#> B2 & C2: "A" 136.125 1 136.125 87.120 <.001 ***
#> Residuals 37.500 24 1.563
#> ────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────────
#> "A" "B" "C" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────────
#> A1 B1 C1 4.000 [ 2.710, 5.290] (0.625)
#> A2 B1 C1 8.000 [ 6.710, 9.290] (0.625)
#> A1 B2 C1 4.000 [ 2.710, 5.290] (0.625)
#> A2 B2 C1 8.000 [ 6.710, 9.290] (0.625)
#> A1 B1 C2 4.750 [ 3.460, 6.040] (0.625)
#> A2 B1 C2 6.000 [ 4.710, 7.290] (0.625)
#> A1 B2 C2 3.750 [ 2.460, 5.040] (0.625)
#> A2 B2 C2 12.000 [10.710, 13.290] (0.625)
#> ────────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ─────────────────────────────────────────────────────────────────────────────
#> Contrast "B" "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ─────────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.741, 4.659]
#> A2 - A1 B2 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.741, 4.659]
#> A2 - A1 B1 C2 1.250 (0.884) 24 1.414 .170 1.000 [-0.459, 2.459]
#> A2 - A1 B2 C2 8.250 (0.884) 24 9.334 <.001 *** 6.600 [ 5.141, 8.059]
#> ─────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.250
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
## Just to name a few...
## You may test other combinations...
#### Within-Subjects Design ####
within.1
#> ID A1 A2 A3 A4
#> 1 S1 3 4 8 9
#> 2 S2 6 6 9 8
#> 3 S3 4 4 8 8
#> 4 S4 3 2 7 7
#> 5 S5 5 4 5 12
#> 6 S6 7 5 6 13
#> 7 S7 5 3 7 12
#> 8 S8 2 3 6 11
MANOVA(within.1, dvs="A1:A4", dvs.pattern="A(.)",
within="A") %>%
EMMEANS("A")
#>
#> Note:
#> dvs="A1:A4" is matched to variables:
#> A1, A2, A3, A4
#>
#> ====== ANOVA (Within-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────
#> "A" Mean S.D. n
#> ─────────────────────
#> A1 4.375 (1.685) 8
#> A2 3.875 (1.246) 8
#> A3 7.000 (1.309) 8
#> A4 10.000 (2.268) 8
#> ─────────────────────
#> Total sample size: N = 8
#>
#> ANOVA Table:
#> Dependent variable(s): A1, A2, A3, A4
#> Between-subjects factor(s): –
#> Within-subjects factor(s): A
#> Covariate(s): –
#> ─────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ─────────────────────────────────────────────────────────────────
#> A 63.375 2.518 3 21 25.170 <.001 *** .782 [.609, .858] .707
#> ─────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> No between-subjects factors. No need to do the Levene’s test.
#>
#> Mauchly’s Test of Sphericity:
#> ────────────────────────
#> Mauchly's W p
#> ────────────────────────
#> A 0.1899 .095 .
#> ────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────
#> Effect df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────
#> A 3 7 47.960 <.001 *** .954 [.848, .977]
#> ────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ───────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────
#> Mean: "A" 0.954 3.000 5.000 34.257 <.001 ***
#> ───────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────
#> "A" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────
#> A1 4.375 [2.966, 5.784] (0.596)
#> A2 3.875 [2.833, 4.917] (0.441)
#> A3 7.000 [5.905, 8.095] (0.463)
#> A4 10.000 [8.104, 11.896] (0.802)
#> ───────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────
#> Contrast Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────
#> A2 - A1 -0.500 (0.423) 7 -1.183 1.000 -0.223 [-0.907, 0.462]
#> A3 - A1 2.625 (0.754) 7 3.479 .062 . 1.170 [-0.053, 2.392]
#> A3 - A2 3.125 (0.515) 7 6.063 .003 ** 1.393 [ 0.558, 2.228]
#> A4 - A1 5.625 (0.778) 7 7.232 .001 ** 2.507 [ 1.247, 3.767]
#> A4 - A2 6.125 (0.875) 7 7.000 .001 ** 2.729 [ 1.312, 4.147]
#> A4 - A3 3.000 (1.180) 7 2.542 .231 1.337 [-0.575, 3.249]
#> ──────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 2.244
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
within.2
#> ID A1B1 A1B2 A1B3 A2B1 A2B2 A2B3
#> 1 S1 3 4 5 4 8 12
#> 2 S2 6 6 7 5 9 13
#> 3 S3 4 4 5 3 8 12
#> 4 S4 3 2 2 3 7 11
MANOVA(within.2, dvs="A1B1:A2B3", dvs.pattern="A(.)B(.)",
within=c("A", "B")) %>%
EMMEANS("A", by="B") %>%
EMMEANS("B", by="A") # singular error matrix
#>
#> Note:
#> dvs="A1B1:A2B3" is matched to variables:
#> A1B1, A1B2, A1B3, A2B1, A2B2, A2B3
#>
#> ====== ANOVA (Within-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────
#> "A" "B" Mean S.D. n
#> ─────────────────────────
#> A1 B1 4.000 (1.414) 4
#> A1 B2 4.000 (1.633) 4
#> A1 B3 4.750 (2.062) 4
#> A2 B1 3.750 (0.957) 4
#> A2 B2 8.000 (0.816) 4
#> A2 B3 12.000 (0.816) 4
#> ─────────────────────────
#> Total sample size: N = 4
#>
#> ANOVA Table:
#> Dependent variable(s): A1B1, A1B2, A1B3, A2B1, A2B2, A2B3
#> Between-subjects factor(s): –
#> Within-subjects factor(s): A, B
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────
#> A 80.667 1.111 1 3 72.600 .003 ** .960 [.699, .985] .707
#> B 40.542 0.264 2 6 153.632 <.001 *** .981 [.930, .991] .708
#> A * B 28.292 0.236 2 6 119.824 <.001 *** .976 [.911, .988] .628
#> ──────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> No between-subjects factors. No need to do the Levene’s test.
#>
#> Mauchly’s Test of Sphericity:
#> ────────────────────────────
#> Mauchly's W p
#> ────────────────────────────
#> B 0.0665 .066 .
#> A * B 0.2491 .249
#> ────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────
#> A B1 1 3 0.273 .638 .083 [.000, .597]
#> A B2 1 3 96.000 .002 ** .970 [.763, .988]
#> A B3 1 3 132.789 .001 ** .978 [.823, .992]
#> ─────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.083 1.000 3.000 0.273 .638
#> B2: "A" 0.970 1.000 3.000 96.000 .002 **
#> B3: "A" 0.978 1.000 3.000 132.789 .001 **
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 1.750, 6.250] (0.707)
#> A2 B1 3.750 [ 2.227, 5.273] (0.479)
#> A1 B2 4.000 [ 1.402, 6.598] (0.816)
#> A2 B2 8.000 [ 6.701, 9.299] (0.408)
#> A1 B3 4.750 [ 1.470, 8.030] (1.031)
#> A2 B3 12.000 [10.701, 13.299] (0.408)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 -0.250 (0.479) 3 -0.522 .638 -0.272 [-1.930, 1.386]
#> A2 - A1 B2 4.000 (0.408) 3 9.798 .002 ** 4.353 [ 2.939, 5.767]
#> A2 - A1 B3 7.250 (0.629) 3 11.523 .001 ** 7.890 [ 5.711, 10.068]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.919
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "B") ------
#>
#> Error in solve.default(zcov, z) :
#> system is computationally singular: reciprocal condition number = 2.23113e-17
#> Error in solve.default(zcov, z) :
#> system is computationally singular: reciprocal condition number = 2.23113e-17
#> Warning: Some CIs could not be estimated due to non-finite F, df, or df_error
#> values.
#> Joint Tests of "B":
#> ───────────────────────────────────────────────────────────────
#> Effect "A" df1 df2 F p η²p [90% CI of η²p]
#> ───────────────────────────────────────────────────────────────
#> B A1 2 3 13.500 .032 * .900 [.219, .962]
#> B A2 2 NA [ NA, NA]
#> (confounded) A2 2 NA [ NA, NA]
#> ───────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Estimated Marginal Means of "B":
#> ────────────────────────────────────────
#> "B" "A" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> B1 A1 4.000 [ 1.750, 6.250] (0.707)
#> B2 A1 4.000 [ 1.402, 6.598] (0.816)
#> B3 A1 4.750 [ 1.470, 8.030] (1.031)
#> B1 A2 3.750 [ 2.227, 5.273] (0.479)
#> B2 A2 8.000 [ 6.701, 9.299] (0.408)
#> B3 A2 12.000 [10.701, 13.299] (0.408)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "B":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "A" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> B2 - B1 A1 0.000 (0.408) 3 0.000 1.000 0.000 [-2.158, 2.158]
#> B3 - B1 A1 0.750 (0.629) 3 1.192 .957 0.816 [-2.509, 4.141]
#> B3 - B2 A1 0.750 (0.250) 3 3.000 .173 0.816 [-0.505, 2.137]
#> B2 - B1 A2 4.250 (0.250) 3 17.000 .001 ** 4.625 [ 3.304, 5.946]
#> B3 - B1 A2 8.250 (0.250) 3 33.000 <.001 *** 8.978 [ 7.656, 10.299]
#> B3 - B2 A2 4.000 (0.000) 3 Inf <.001 *** 4.353 [ 4.353, 4.353]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.919
#> P-value adjustment: Bonferroni method for 3 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
# :::::::::::::::::::::::::::::::::::::::
# This would produce a WARNING because of
# the linear dependence of A2B2 and A2B3.
# See: Corr(within.2[c("A2B2", "A2B3")])
within.3
#> ID A1B1C1 A1B1C2 A1B2C1 A1B2C2 A2B1C1 A2B1C2 A2B2C1 A2B2C2
#> 1 S1 3 5 4 4 8 5 9 12
#> 2 S2 6 7 6 5 9 6 8 13
#> 3 S3 4 5 4 3 8 7 8 12
#> 4 S4 3 2 2 3 7 6 7 11
MANOVA(within.3, dvs="A1B1C1:A2B2C2", dvs.pattern="A(.)B(.)C(.)",
within=c("A", "B", "C")) %>%
EMMEANS("A", by="B") %>%
EMMEANS(c("A", "B"), by="C") %>%
EMMEANS("A", by=c("B", "C"))
#>
#> Note:
#> dvs="A1B1C1:A2B2C2" is matched to variables:
#> A1B1C1, A1B1C2, A1B2C1, A1B2C2, A2B1C1, A2B1C2, A2B2C1, A2B2C2
#>
#> ====== ANOVA (Within-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────────
#> "A" "B" "C" Mean S.D. n
#> ─────────────────────────────
#> A1 B1 C1 4.000 (1.414) 4
#> A1 B1 C2 4.750 (2.062) 4
#> A1 B2 C1 4.000 (1.633) 4
#> A1 B2 C2 3.750 (0.957) 4
#> A2 B1 C1 8.000 (0.816) 4
#> A2 B1 C2 6.000 (0.816) 4
#> A2 B2 C1 8.000 (0.816) 4
#> A2 B2 C2 12.000 (0.816) 4
#> ─────────────────────────────
#> Total sample size: N = 4
#>
#> ANOVA Table:
#> Dependent variable(s): A1B1C1, A1B1C2, A1B2C1, A1B2C2, A2B1C1, A2B1C2, A2B2C1, A2B2C2
#> Between-subjects factor(s): –
#> Within-subjects factor(s): A, B, C
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 153.125 1.875 1 3 81.667 .003 ** .965 [.727, .986] .803
#> B 12.500 0.583 1 3 21.429 .019 * .877 [.279, .954] .250
#> C 3.125 0.042 1 3 75.000 .003 ** .962 [.707, .985] .077
#> A * B 24.500 0.250 1 3 98.000 .002 ** .970 [.768, .989] .395
#> A * C 1.125 0.708 1 3 1.588 .297 .346 [.000, .751] .029
#> B * C 12.500 0.417 1 3 30.000 .012 * .909 [.411, .965] .250
#> A * B * C 24.500 1.083 1 3 22.615 .018 * .883 [.300, .956] .395
#> ──────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> No between-subjects factors. No need to do the Levene’s test.
#>
#> Mauchly’s Test of Sphericity:
#> The repeated measures have only two levels. The assumption of sphericity is always met.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────
#> C B1 1 3 6.818 .080 . .694 [.000, .886]
#> C B2 1 3 61.364 .004 ** .953 [.653, .982]
#> A B1 1 3 17.640 .025 * .855 [.202, .945]
#> A B2 1 3 266.778 <.001 *** .989 [.908, .996]
#> C * A B1 1 3 6.153 .089 . .672 [.000, .878]
#> C * A B2 1 3 32.111 .011 * .915 [.436, .968]
#> ─────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.855 1.000 3.000 17.640 .025 *
#> B2: "A" 0.989 1.000 3.000 266.778 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────
#> A1 B1 4.375 [1.746, 7.004] (0.826)
#> A2 B1 7.000 [6.081, 7.919] (0.289)
#> A1 B2 3.875 [1.886, 5.864] (0.625)
#> A2 B2 10.000 [8.875, 11.125] (0.354)
#> ───────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 2.625 (0.625) 3 4.200 .025 * 2.205 [0.534, 3.877]
#> A2 - A1 B2 6.125 (0.375) 3 16.333 <.001 *** 5.146 [4.143, 6.149]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.190
#> Results are averaged over the levels of: C
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A" & "B") ------
#>
#> Error in solve.default(zcov, z) :
#> system is computationally singular: reciprocal condition number = 1.67575e-17
#> Error in solve.default(zcov, z) :
#> system is computationally singular: reciprocal condition number = 1.38778e-17
#> Joint Tests of "A" & "B":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> B C1 1 3 0.000 1.000 .000 [.000, .000]
#> B C2 1 3 50.000 .006 ** .943 [.592, .978]
#> A C1 1 3 54.857 .005 ** .948 [.620, .980]
#> A C2 1 3 63.706 .004 ** .955 [.664, .983]
#> B * A C1 1 3 0.000 1.000 .000 [.000, .000]
#> B * A C2 1 3 42.000 .007 ** .933 [.534, .975]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A" & "B":
#> ───────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────────
#> C1: "A" & "B" 0.000 1.000 3.000 0.000 1.000
#> C2: "A" & "B" 0.933 1.000 3.000 42.000 .007 **
#> ───────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A" & "B":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 1.750, 6.250] (0.707)
#> A2 B1 8.000 [ 6.701, 9.299] (0.408)
#> A1 B2 4.000 [ 1.402, 6.598] (0.816)
#> A2 B2 8.000 [ 6.701, 9.299] (0.408)
#> A1 B1 4.750 [ 1.470, 8.030] (1.031)
#> A2 B1 6.000 [ 4.701, 7.299] (0.408)
#> A1 B2 3.750 [ 2.227, 5.273] (0.479)
#> A2 B2 12.000 [10.701, 13.299] (0.408)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A" & "B":
#> ───────────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ───────────────────────────────────────────────────────────────────────────────
#> A2 B1 - A1 B1 C1 4.000 (0.408) 3 9.798 .014 * 3.361 [ 1.223, 5.498]
#> A1 B2 - A1 B1 C1 0.000 (0.408) 3 0.000 1.000 0.000 [-2.137, 2.137]
#> A1 B2 - A2 B1 C1 -4.000 (0.408) 3 -9.798 .014 * -3.361 [-5.498, -1.223]
#> A2 B2 - A1 B1 C1 4.000 (0.816) 3 4.899 .098 . 3.361 [-0.914, 7.635]
#> A2 B2 - A2 B1 C1 0.000 (0.408) 3 0.000 1.000 0.000 [-2.137, 2.137]
#> A2 B2 - A1 B2 C1 4.000 (0.707) 3 5.657 .066 . 3.361 [-0.341, 7.063]
#> A2 B1 - A1 B1 C2 1.250 (1.109) 3 1.127 1.000 1.050 [-4.754, 6.855]
#> A1 B2 - A1 B1 C2 -1.000 (0.707) 3 -1.414 1.000 -0.840 [-4.542, 2.862]
#> A1 B2 - A2 B1 C2 -2.250 (0.750) 3 -3.000 .346 -1.890 [-5.817, 2.036]
#> A2 B2 - A1 B1 C2 7.250 (0.629) 3 11.523 .008 ** 6.091 [ 2.797, 9.385]
#> A2 B2 - A2 B1 C2 6.000 (0.577) 3 10.392 .011 * 5.041 [ 2.018, 8.064]
#> A2 B2 - A1 B2 C2 8.250 (0.250) 3 33.000 <.001 *** 6.931 [ 5.623, 8.240]
#> ───────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.190
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ──────────────────────────────────────────────────────────────
#> Effect "B" "C" df1 df2 F p η²p [90% CI of η²p]
#> ──────────────────────────────────────────────────────────────
#> A B1 C1 1 3 96.000 .002 ** .970 [.763, .988]
#> A B2 C1 1 3 32.000 .011 * .914 [.435, .967]
#> A B1 C2 1 3 1.271 .342 .298 [.000, .729]
#> A B2 C2 1 3 1089.000 <.001 *** .997 [.977, .999]
#> ──────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ───────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────────
#> B1 & C1: "A" 0.970 1.000 3.000 96.000 .002 **
#> B2 & C1: "A" 0.914 1.000 3.000 32.000 .011 *
#> B1 & C2: "A" 0.298 1.000 3.000 1.271 .342
#> B2 & C2: "A" 0.997 1.000 3.000 1089.000 <.001 ***
#> ───────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────────
#> "A" "B" "C" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────────
#> A1 B1 C1 4.000 [ 1.750, 6.250] (0.707)
#> A2 B1 C1 8.000 [ 6.701, 9.299] (0.408)
#> A1 B2 C1 4.000 [ 1.402, 6.598] (0.816)
#> A2 B2 C1 8.000 [ 6.701, 9.299] (0.408)
#> A1 B1 C2 4.750 [ 1.470, 8.030] (1.031)
#> A2 B1 C2 6.000 [ 4.701, 7.299] (0.408)
#> A1 B2 C2 3.750 [ 2.227, 5.273] (0.479)
#> A2 B2 C2 12.000 [10.701, 13.299] (0.408)
#> ────────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────────
#> Contrast "B" "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 C1 4.000 (0.408) 3 9.798 .002 ** 3.361 [ 2.269, 4.452]
#> A2 - A1 B2 C1 4.000 (0.707) 3 5.657 .011 * 3.361 [ 1.470, 5.251]
#> A2 - A1 B1 C2 1.250 (1.109) 3 1.127 .342 1.050 [-1.914, 4.015]
#> A2 - A1 B2 C2 8.250 (0.250) 3 33.000 <.001 *** 6.931 [ 6.263, 7.600]
#> ──────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.190
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
## Just to name a few...
## You may test other combinations...
#### Mixed Design ####
mixed.2_1b1w
#> A B1 B2 B3
#> 1 1 3 4 5
#> 2 1 6 6 7
#> 3 1 4 4 5
#> 4 1 3 2 2
#> 5 2 4 8 12
#> 6 2 5 9 13
#> 7 2 3 8 12
#> 8 2 3 7 11
MANOVA(mixed.2_1b1w, dvs="B1:B3", dvs.pattern="B(.)",
between="A", within="B", sph.correction="GG") %>%
EMMEANS("A", by="B") %>%
EMMEANS("B", by="A")
#>
#> Note:
#> dvs="B1:B3" is matched to variables:
#> B1, B2, B3
#>
#> ====== ANOVA (Mixed Design) ======
#>
#> Descriptives:
#> ─────────────────────────
#> "A" "B" Mean S.D. n
#> ─────────────────────────
#> A1 B1 4.000 (1.414) 4
#> A1 B2 4.000 (1.633) 4
#> A1 B3 4.750 (2.062) 4
#> A2 B1 3.750 (0.957) 4
#> A2 B2 8.000 (0.816) 4
#> A2 B3 12.000 (0.816) 4
#> ─────────────────────────
#> Total sample size: N = 8
#>
#> ANOVA Table:
#> Dependent variable(s): B1, B2, B3
#> Between-subjects factor(s): A
#> Within-subjects factor(s): B
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 80.667 5.083 1.000 6.000 15.869 .007 ** .726 [.248, .871] .707
#> B 74.702 0.461 1.085 6.513 162.167 <.001 *** .964 [.880, .983] .708
#> A * B 52.130 0.461 1.085 6.513 113.167 <.001 *** .950 [.833, .976] .628
#> ──────────────────────────────────────────────────────────────────────────
#> Sphericity correction method: GG (Greenhouse-Geisser)
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ────────────────────────────────────
#> Levene’s F df1 df2 p
#> ────────────────────────────────────
#> DV: B1 0.300 1 6 .604
#> DV: B2 0.600 1 6 .468
#> DV: B3 1.485 1 6 .269
#> ────────────────────────────────────
#>
#> Mauchly’s Test of Sphericity:
#> ────────────────────────────
#> Mauchly's W p
#> ────────────────────────────
#> B 0.1574 .010 **
#> A * B 0.1574 .010 **
#> ────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A B1 1 6 0.086 .780 .014 [.000, .339]
#> A B2 1 6 19.200 .005 ** .762 [.314, .888]
#> A B3 1 6 42.763 <.001 *** .877 [.593, .942]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.014 1.000 6.000 0.086 .780
#> B2: "A" 0.762 1.000 6.000 19.200 .005 **
#> B3: "A" 0.877 1.000 6.000 42.763 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 2.523, 5.477] (0.604)
#> A2 B1 3.750 [ 2.273, 5.227] (0.604)
#> A1 B2 4.000 [ 2.421, 5.579] (0.645)
#> A2 B2 8.000 [ 6.421, 9.579] (0.645)
#> A1 B3 4.750 [ 2.832, 6.668] (0.784)
#> A2 B3 12.000 [10.082, 13.918] (0.784)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 -0.250 (0.854) 6 -0.293 .780 -0.382 [-3.574, 2.810]
#> A2 - A1 B2 4.000 (0.913) 6 4.382 .005 ** 6.110 [ 2.698, 9.522]
#> A2 - A1 B3 7.250 (1.109) 6 6.539 <.001 *** 11.075 [ 6.931, 15.218]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.655
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "B") ------
#>
#> Joint Tests of "B":
#> ─────────────────────────────────────────────────────────
#> Effect "A" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────
#> B A1 2 6 17.471 .003 ** .853 [.492, .930]
#> B A2 2 6 265.941 <.001 *** .989 [.959, .995]
#> ─────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "B":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> A1: "B" 0.853 2.000 5.000 14.559 .008 **
#> A2: "B" 0.989 2.000 5.000 221.618 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "B":
#> ────────────────────────────────────────
#> "B" "A" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> B1 A1 4.000 [ 2.523, 5.477] (0.604)
#> B2 A1 4.000 [ 2.421, 5.579] (0.645)
#> B3 A1 4.750 [ 2.832, 6.668] (0.784)
#> B1 A2 3.750 [ 2.273, 5.227] (0.604)
#> B2 A2 8.000 [ 6.421, 9.579] (0.645)
#> B3 A2 12.000 [10.082, 13.918] (0.784)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "B":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "A" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> B2 - B1 A1 -0.000 (0.339) 6 -0.000 1.000 -0.000 [-1.700, 1.700]
#> B3 - B1 A1 0.750 (0.479) 6 1.567 .505 1.146 [-1.258, 3.550]
#> B3 - B2 A1 0.750 (0.177) 6 4.243 .016 * 1.146 [ 0.258, 2.033]
#> B2 - B1 A2 4.250 (0.339) 6 12.555 <.001 *** 6.492 [ 4.792, 8.192]
#> B3 - B1 A2 8.250 (0.479) 6 17.234 <.001 *** 12.602 [10.198, 15.006]
#> B3 - B2 A2 4.000 (0.177) 6 22.627 <.001 *** 6.110 [ 5.222, 6.998]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.655
#> P-value adjustment: Bonferroni method for 3 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
mixed.3_1b2w
#> A B1C1 B1C2 B2C1 B2C2
#> 1 1 3 5 4 4
#> 2 1 6 7 6 5
#> 3 1 4 5 4 3
#> 4 1 3 2 2 3
#> 5 2 8 5 9 12
#> 6 2 9 6 8 13
#> 7 2 8 7 8 12
#> 8 2 7 6 7 11
MANOVA(mixed.3_1b2w, dvs="B1C1:B2C2", dvs.pattern="B(.)C(.)",
between="A", within=c("B", "C")) %>%
EMMEANS("A", by="B") %>%
EMMEANS(c("A", "B"), by="C") %>%
EMMEANS("A", by=c("B", "C"))
#>
#> Note:
#> dvs="B1C1:B2C2" is matched to variables:
#> B1C1, B1C2, B2C1, B2C2
#>
#> ====== ANOVA (Mixed Design) ======
#>
#> Descriptives:
#> ─────────────────────────────
#> "A" "B" "C" Mean S.D. n
#> ─────────────────────────────
#> A1 B1 C1 4.000 (1.414) 4
#> A1 B1 C2 4.750 (2.062) 4
#> A1 B2 C1 4.000 (1.633) 4
#> A1 B2 C2 3.750 (0.957) 4
#> A2 B1 C1 8.000 (0.816) 4
#> A2 B1 C2 6.000 (0.816) 4
#> A2 B2 C1 8.000 (0.816) 4
#> A2 B2 C2 12.000 (0.816) 4
#> ─────────────────────────────
#> Total sample size: N = 8
#>
#> ANOVA Table:
#> Dependent variable(s): B1C1, B1C2, B2C1, B2C2
#> Between-subjects factor(s): A
#> Within-subjects factor(s): B, C
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 153.125 4.708 1 6 32.522 .001 ** .844 [.503, .926] .803
#> B 12.500 0.417 1 6 30.000 .002 ** .833 [.475, .921] .250
#> A * B 24.500 0.417 1 6 58.800 <.001 *** .907 [.684, .956] .395
#> C 3.125 0.375 1 6 8.333 .028 * .581 [.064, .801] .077
#> A * C 1.125 0.375 1 6 3.000 .134 .333 [.000, .671] .029
#> B * C 12.500 0.750 1 6 16.667 .006 ** .735 [.264, .875] .250
#> A * B * C 24.500 0.750 1 6 32.667 .001 ** .845 [.505, .927] .395
#> ──────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ──────────────────────────────────────
#> Levene’s F df1 df2 p
#> ──────────────────────────────────────
#> DV: B1C1 1.000 1 6 .356
#> DV: B1C2 1.485 1 6 .269
#> DV: B2C1 0.600 1 6 .468
#> DV: B2C2 0.500 1 6 .506
#> ──────────────────────────────────────
#>
#> Mauchly’s Test of Sphericity:
#> The repeated measures have only two levels. The assumption of sphericity is always met.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A B1 1 6 9.000 .024 * .600 [.082, .810]
#> A B2 1 6 72.758 <.001 *** .924 [.736, .964]
#> C B1 1 6 2.143 .194 .263 [.000, .629]
#> C B2 1 6 35.526 <.001 *** .856 [.533, .932]
#> A * C B1 1 6 10.371 .018 * .633 [.117, .827]
#> A * C B2 1 6 45.632 <.001 *** .884 [.613, .945]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.600 1.000 6.000 9.000 .024 *
#> B2: "A" 0.924 1.000 6.000 72.758 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────
#> A1 B1 4.375 [2.861, 5.889] (0.619)
#> A2 B1 7.000 [5.486, 8.514] (0.619)
#> A1 B2 3.875 [2.633, 5.117] (0.508)
#> A2 B2 10.000 [8.758, 11.242] (0.508)
#> ───────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ─────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ─────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 2.625 (0.875) 6 3.000 .024 * 2.797 [0.516, 5.078]
#> A2 - A1 B2 6.125 (0.718) 6 8.530 <.001 *** 6.526 [4.654, 8.398]
#> ─────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.939
#> Results are averaged over the levels of: C
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A" & "B") ------
#>
#> Joint Tests of "A" & "B":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A C1 1 6 24.000 .003 ** .800 [.395, .906]
#> A C2 1 6 37.345 <.001 *** .862 [.550, .935]
#> B C1 1 6 0.000 1.000 .000 [.000, .000]
#> B C2 1 6 30.000 .002 ** .833 [.475, .921]
#> A * B C1 1 6 0.000 1.000 .000 [.000, .000]
#> A * B C2 1 6 58.800 <.001 *** .907 [.684, .956]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A" & "B":
#> ───────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────────
#> C1: "A" & "B" 0.000 1.000 6.000 0.000 1.000
#> C2: "A" & "B" 0.907 1.000 6.000 58.800 <.001 ***
#> ───────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A" & "B":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 2.587, 5.413] (0.577)
#> A2 B1 8.000 [ 6.587, 9.413] (0.577)
#> A1 B2 4.000 [ 2.421, 5.579] (0.645)
#> A2 B2 8.000 [ 6.421, 9.579] (0.645)
#> A1 B1 4.750 [ 2.832, 6.668] (0.784)
#> A2 B1 6.000 [ 4.082, 7.918] (0.784)
#> A1 B2 3.750 [ 2.661, 4.839] (0.445)
#> A2 B2 12.000 [10.911, 13.089] (0.445)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A" & "B":
#> ───────────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ───────────────────────────────────────────────────────────────────────────────
#> A2 B1 - A1 B1 C1 4.000 (0.816) 6 4.899 .016 * 4.262 [ 0.901, 7.622]
#> A1 B2 - A1 B1 C1 0.000 (0.408) 6 0.000 1.000 0.000 [-1.680, 1.680]
#> A1 B2 - A2 B1 C1 -4.000 (0.866) 6 -4.619 .022 * -4.262 [-7.826, -0.697]
#> A2 B2 - A1 B1 C1 4.000 (0.866) 6 4.619 .022 * 4.262 [ 0.697, 7.826]
#> A2 B2 - A2 B1 C1 0.000 (0.408) 6 0.000 1.000 0.000 [-1.680, 1.680]
#> A2 B2 - A1 B2 C1 4.000 (0.913) 6 4.382 .028 * 4.262 [ 0.505, 8.019]
#> A2 B1 - A1 B1 C2 1.250 (1.109) 6 1.127 1.000 1.332 [-3.231, 5.895]
#> A1 B2 - A1 B1 C2 -1.000 (0.645) 6 -1.549 1.000 -1.065 [-3.722, 1.591]
#> A1 B2 - A2 B1 C2 -2.250 (0.901) 6 -2.496 .281 -2.397 [-6.107, 1.313]
#> A2 B2 - A1 B1 C2 7.250 (0.901) 6 8.043 .001 ** 7.724 [ 4.014, 11.434]
#> A2 B2 - A2 B1 C2 6.000 (0.645) 6 9.295 <.001 *** 6.393 [ 3.736, 9.049]
#> A2 B2 - A1 B2 C2 8.250 (0.629) 6 13.113 <.001 *** 8.790 [ 6.200, 11.379]
#> ───────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.939
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Effect "B" "C" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────────
#> A B1 C1 1 6 24.000 .003 ** .800 [.395, .906]
#> A B2 C1 1 6 19.200 .005 ** .762 [.314, .888]
#> A B1 C2 1 6 1.271 .303 .175 [.000, .568]
#> A B2 C2 1 6 171.947 <.001 *** .966 [.879, .984]
#> ─────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ──────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ──────────────────────────────────────────────────────────────────
#> B1 & C1: "A" 0.800 1.000 6.000 24.000 .003 **
#> B2 & C1: "A" 0.762 1.000 6.000 19.200 .005 **
#> B1 & C2: "A" 0.175 1.000 6.000 1.271 .303
#> B2 & C2: "A" 0.966 1.000 6.000 171.947 <.001 ***
#> ──────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────────
#> "A" "B" "C" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────────
#> A1 B1 C1 4.000 [ 2.587, 5.413] (0.577)
#> A2 B1 C1 8.000 [ 6.587, 9.413] (0.577)
#> A1 B2 C1 4.000 [ 2.421, 5.579] (0.645)
#> A2 B2 C1 8.000 [ 6.421, 9.579] (0.645)
#> A1 B1 C2 4.750 [ 2.832, 6.668] (0.784)
#> A2 B1 C2 6.000 [ 4.082, 7.918] (0.784)
#> A1 B2 C2 3.750 [ 2.661, 4.839] (0.445)
#> A2 B2 C2 12.000 [10.911, 13.089] (0.445)
#> ────────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────────
#> Contrast "B" "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 C1 4.000 (0.816) 6 4.899 .003 ** 4.262 [ 2.133, 6.390]
#> A2 - A1 B2 C1 4.000 (0.913) 6 4.382 .005 ** 4.262 [ 1.882, 6.642]
#> A2 - A1 B1 C2 1.250 (1.109) 6 1.127 .303 1.332 [-1.559, 4.222]
#> A2 - A1 B2 C2 8.250 (0.629) 6 13.113 <.001 *** 8.790 [ 7.150, 10.430]
#> ──────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.939
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
## Just to name a few...
## You may test other combinations...
mixed.3_2b1w
#> A C B1 B2
#> 1 1 1 3 4
#> 2 1 1 6 6
#> 3 1 1 4 4
#> 4 1 1 3 2
#> 5 1 2 5 4
#> 6 1 2 7 5
#> 7 1 2 5 3
#> 8 1 2 2 3
#> 9 2 1 8 9
#> 10 2 1 9 8
#> 11 2 1 8 8
#> 12 2 1 7 7
#> 13 2 2 5 12
#> 14 2 2 6 13
#> 15 2 2 7 12
#> 16 2 2 6 11
MANOVA(mixed.3_2b1w, dvs="B1:B2", dvs.pattern="B(.)",
between=c("A", "C"), within="B") %>%
EMMEANS("A", by="B") %>%
EMMEANS("A", by="C") %>%
EMMEANS(c("A", "B"), by="C") %>%
EMMEANS("B", by=c("A", "C"))
#>
#> Note:
#> dvs="B1:B2" is matched to variables:
#> B1, B2
#>
#> ====== ANOVA (Mixed Design) ======
#>
#> Descriptives:
#> ─────────────────────────────
#> "A" "C" "B" Mean S.D. n
#> ─────────────────────────────
#> A1 C1 B1 4.000 (1.414) 4
#> A1 C1 B2 4.000 (1.633) 4
#> A1 C2 B1 4.750 (2.062) 4
#> A1 C2 B2 3.750 (0.957) 4
#> A2 C1 B1 8.000 (0.816) 4
#> A2 C1 B2 8.000 (0.816) 4
#> A2 C2 B1 6.000 (0.816) 4
#> A2 C2 B2 12.000 (0.816) 4
#> ─────────────────────────────
#> Total sample size: N = 16
#>
#> ANOVA Table:
#> Dependent variable(s): B1, B2
#> Between-subjects factor(s): A, C
#> Within-subjects factor(s): B
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 153.125 2.542 1 12 60.246 <.001 *** .834 [.639, .906] .803
#> C 3.125 2.542 1 12 1.230 .289 .093 [.000, .390] .077
#> A * C 1.125 2.542 1 12 0.443 .518 .036 [.000, .305] .029
#> B 12.500 0.583 1 12 21.429 <.001 *** .641 [.308, .795] .250
#> A * B 24.500 0.583 1 12 42.000 <.001 *** .778 [.532, .874] .395
#> C * B 12.500 0.583 1 12 21.429 <.001 *** .641 [.308, .795] .250
#> A * C * B 24.500 0.583 1 12 42.000 <.001 *** .778 [.532, .874] .395
#> ──────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ────────────────────────────────────
#> Levene’s F df1 df2 p
#> ────────────────────────────────────
#> DV: B1 0.946 3 12 .449
#> DV: B2 0.423 3 12 .740
#> ────────────────────────────────────
#>
#> Mauchly’s Test of Sphericity:
#> The repeated measures have only two levels. The assumption of sphericity is always met.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────
#> A B1 1 12 14.538 .002 ** .548 [.190, .739]
#> A B2 1 12 122.085 <.001 *** .911 [.799, .949]
#> C B1 1 12 0.824 .382 .064 [.000, .353]
#> C B2 1 12 11.441 .005 ** .488 [.129, .702]
#> A * C B1 1 12 3.989 .069 . .249 [.000, .537]
#> A * C B2 1 12 14.695 .002 ** .550 [.193, .741]
#> ─────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.548 1.000 12.000 14.538 .002 **
#> B2: "A" 0.911 1.000 12.000 122.085 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────
#> A1 B1 4.375 [3.314, 5.436] (0.487)
#> A2 B1 7.000 [5.939, 8.061] (0.487)
#> A1 B2 3.875 [3.021, 4.729] (0.392)
#> A2 B2 10.000 [9.146, 10.854] (0.392)
#> ───────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 2.625 (0.688) 12 3.813 .002 ** 2.717 [1.164, 4.270]
#> A2 - A1 B2 6.125 (0.554) 12 11.049 <.001 *** 6.340 [5.090, 7.590]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.966
#> Results are averaged over the levels of: C
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A C1 1 12 25.180 <.001 *** .677 [.361, .816]
#> A C2 1 12 35.508 <.001 *** .747 [.477, .856]
#> B C1 1 12 0.000 1.000 .000 [.000, .000]
#> B C2 1 12 42.857 <.001 *** .781 [.538, .876]
#> A * B C1 1 12 0.000 1.000 .000 [.000, .000]
#> A * B C2 1 12 84.000 <.001 *** .875 [.724, .929]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> C1: "A" 0.677 1.000 12.000 25.180 <.001 ***
#> C2: "A" 0.747 1.000 12.000 35.508 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ──────────────────────────────────────
#> "A" "C" Mean [95% CI of Mean] S.E.
#> ──────────────────────────────────────
#> A1 C1 4.000 [2.772, 5.228] (0.564)
#> A2 C1 8.000 [6.772, 9.228] (0.564)
#> A1 C2 4.250 [3.022, 5.478] (0.564)
#> A2 C2 9.000 [7.772, 10.228] (0.564)
#> ──────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ─────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ─────────────────────────────────────────────────────────────────────────
#> A2 - A1 C1 4.000 (0.797) 12 5.018 <.001 *** 4.140 [2.343, 5.938]
#> A2 - A1 C2 4.750 (0.797) 12 5.959 <.001 *** 4.917 [3.119, 6.714]
#> ─────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.966
#> Results are averaged over the levels of: B
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A" & "B") ------
#>
#> Joint Tests of "A" & "B":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A C1 1 12 25.180 <.001 *** .677 [.361, .816]
#> A C2 1 12 35.508 <.001 *** .747 [.477, .856]
#> B C1 1 12 0.000 1.000 .000 [.000, .000]
#> B C2 1 12 42.857 <.001 *** .781 [.538, .876]
#> A * B C1 1 12 0.000 1.000 .000 [.000, .000]
#> A * B C2 1 12 84.000 <.001 *** .875 [.724, .929]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A" & "B":
#> ───────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────────
#> C1: "A" & "B" 0.000 1.000 12.000 0.000 1.000
#> C2: "A" & "B" 0.875 1.000 12.000 84.000 <.001 ***
#> ───────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A" & "B":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 2.500, 5.500] (0.688)
#> A2 B1 8.000 [ 6.500, 9.500] (0.688)
#> A1 B2 4.000 [ 2.792, 5.208] (0.554)
#> A2 B2 8.000 [ 6.792, 9.208] (0.554)
#> A1 B1 4.750 [ 3.250, 6.250] (0.688)
#> A2 B1 6.000 [ 4.500, 7.500] (0.688)
#> A1 B2 3.750 [ 2.542, 4.958] (0.554)
#> A2 B2 12.000 [10.792, 13.208] (0.554)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A" & "B":
#> ───────────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ───────────────────────────────────────────────────────────────────────────────
#> A2 B1 - A1 B1 C1 4.000 (0.974) 12 4.108 .009 ** 4.140 [ 0.963, 7.318]
#> A1 B2 - A1 B1 C1 -0.000 (0.540) 12 -0.000 1.000 -0.000 [-1.762, 1.762]
#> A1 B2 - A2 B1 C1 -4.000 (0.884) 12 -4.525 .004 ** -4.140 [-7.025, -1.256]
#> A2 B2 - A1 B1 C1 4.000 (0.884) 12 4.525 .004 ** 4.140 [ 1.256, 7.025]
#> A2 B2 - A2 B1 C1 0.000 (0.540) 12 0.000 1.000 0.000 [-1.762, 1.762]
#> A2 B2 - A1 B2 C1 4.000 (0.784) 12 5.102 .002 ** 4.140 [ 1.582, 6.699]
#> A2 B1 - A1 B1 C2 1.250 (0.974) 12 1.284 1.000 1.294 [-1.883, 4.471]
#> A1 B2 - A1 B1 C2 -1.000 (0.540) 12 -1.852 .533 -1.035 [-2.798, 0.727]
#> A1 B2 - A2 B1 C2 -2.250 (0.884) 12 -2.546 .154 -2.329 [-5.213, 0.555]
#> A2 B2 - A1 B1 C2 7.250 (0.884) 12 8.202 <.001 *** 7.504 [ 4.620, 10.389]
#> A2 B2 - A2 B1 C2 6.000 (0.540) 12 11.110 <.001 *** 6.211 [ 4.448, 7.973]
#> A2 B2 - A1 B2 C2 8.250 (0.784) 12 10.524 <.001 *** 8.540 [ 5.981, 11.098]
#> ───────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.966
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "B") ------
#>
#> Joint Tests of "B":
#> ─────────────────────────────────────────────────────────────
#> Effect "A" "C" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────────
#> B A1 C1 1 12 0.000 1.000 .000 [.000, .000]
#> B A2 C1 1 12 0.000 1.000 .000 [.000, .000]
#> B A1 C2 1 12 3.429 .089 . .222 [.000, .515]
#> B A2 C2 1 12 123.429 <.001 *** .911 [.801, .950]
#> ─────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "B":
#> ──────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ──────────────────────────────────────────────────────────────────
#> A1 & C1: "B" 0.000 1.000 12.000 0.000 1.000
#> A2 & C1: "B" 0.000 1.000 12.000 0.000 1.000
#> A1 & C2: "B" 0.222 1.000 12.000 3.429 .089 .
#> A2 & C2: "B" 0.911 1.000 12.000 123.429 <.001 ***
#> ──────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "B":
#> ────────────────────────────────────────────
#> "B" "A" "C" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────────
#> B1 A1 C1 4.000 [ 2.500, 5.500] (0.688)
#> B2 A1 C1 4.000 [ 2.792, 5.208] (0.554)
#> B1 A2 C1 8.000 [ 6.500, 9.500] (0.688)
#> B2 A2 C1 8.000 [ 6.792, 9.208] (0.554)
#> B1 A1 C2 4.750 [ 3.250, 6.250] (0.688)
#> B2 A1 C2 3.750 [ 2.542, 4.958] (0.554)
#> B1 A2 C2 6.000 [ 4.500, 7.500] (0.688)
#> B2 A2 C2 12.000 [10.792, 13.208] (0.554)
#> ────────────────────────────────────────────
#>
#> Pairwise Comparisons of "B":
#> ──────────────────────────────────────────────────────────────────────────────
#> Contrast "A" "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────────
#> B2 - B1 A1 C1 -0.000 (0.540) 12 -0.000 1.000 -0.000 [-1.218, 1.218]
#> B2 - B1 A2 C1 0.000 (0.540) 12 0.000 1.000 0.000 [-1.218, 1.218]
#> B2 - B1 A1 C2 -1.000 (0.540) 12 -1.852 .089 . -1.035 [-2.253, 0.183]
#> B2 - B1 A2 C2 6.000 (0.540) 12 11.110 <.001 *** 6.211 [ 4.993, 7.429]
#> ──────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.966
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
## Just to name a few...
## You may test other combinations...
#### Other Examples ####
air = airquality
air$Day.1or2 = ifelse(air$Day %% 2 == 1, 1, 2) %>%
factor(levels=1:2, labels=c("odd", "even"))
MANOVA(air, dv="Temp", between=c("Month", "Day.1or2"),
covariate=c("Solar.R", "Wind")) %>%
EMMEANS("Month", contrast="seq") %>%
EMMEANS("Month", by="Day.1or2", contrast="poly")
#> Warning: Numerical variables NOT centered on 0 (matters if variable in interaction):
#> NA, Wind
#> Warning: Missing values for 7 ID(s), which were removed before analysis:
#> 5, 6, 11, 27, 96, 97, 98
#> Below the first few rows (in wide format) of the removed cases with missing data.
#> bruceR.ID Month Day.1or2 Solar.R Wind .
#> # 5 5 Month5 odd NA 14.3 56
#> # 6 6 Month5 even NA 14.9 66
#> # 11 11 Month5 odd NA 6.9 74
#> # 27 27 Month5 odd NA 8.0 57
#> # 96 96 Month8 even NA 6.9 86
#> # 97 97 Month8 odd NA 7.4 85
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────────────────
#> "Month" "Day.1or2" Mean S.D. n
#> ─────────────────────────────────────
#> Month5 odd 66.077 (7.297) 13
#> Month5 even 65.714 (6.390) 14
#> Month6 odd 78.600 (6.905) 15
#> Month6 even 79.600 (6.479) 15
#> Month7 odd 83.375 (4.064) 16
#> Month7 even 84.467 (4.642) 15
#> Month8 odd 83.667 (6.102) 15
#> Month8 even 83.846 (7.978) 13
#> Month9 odd 76.733 (9.177) 15
#> Month9 even 77.067 (7.769) 15
#> ─────────────────────────────────────
#> Total sample size: N = 153 (7 missing observations deleted)
#>
#> ANOVA Table:
#> Dependent variable(s): Temp
#> Between-subjects factor(s): Month, Day.1or2
#> Within-subjects factor(s): –
#> Covariate(s): Solar.R, Wind
#> ───────────────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ───────────────────────────────────────────────────────────────────────────────────
#> Month 1059.183 35.312 4 134 29.995 <.001 *** .472 [.366, .550] .472
#> Day.1or2 6.804 35.312 1 134 0.193 .661 .001 [.000, .030] .001
#> Solar.R 612.990 35.312 1 134 17.359 <.001 *** .115 [.043, .204] .115
#> Wind 880.158 35.312 1 134 24.925 <.001 *** .157 [.074, .251] .157
#> Month * Day.1or2 7.174 35.312 4 134 0.203 .936 .006 [.000, .004] .006
#> ───────────────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ──────────────────────────────────────
#> Levene’s F df1 df2 p
#> ──────────────────────────────────────
#> DV: Temp 1.573 9 143 .129
#> ──────────────────────────────────────
#>
#> ------ EMMEANS (effect = "Month") ------
#>
#> Joint Tests of "Month":
#> ──────────────────────────────────────────────────────────────
#> Effect df1 df2 F p η²p [90% CI of η²p]
#> ──────────────────────────────────────────────────────────────
#> Month 4 134 29.995 <.001 *** .472 [.366, .550]
#> Day.1or2 1 134 0.193 .661 .001 [.000, .030]
#> Solar.R 1 134 17.359 <.001 *** .115 [.043, .204]
#> Wind 1 134 24.925 <.001 *** .157 [.074, .251]
#> Month * Day.1or2 4 134 0.203 .936 .006 [.000, .004]
#> ──────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "Month":
#> ──────────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ──────────────────────────────────────────────────────────────
#> Mean: "Month" 4236.730 4 1059.183 29.995 <.001 ***
#> Residuals 4731.762 134 35.312
#> ──────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "Month":
#> ────────────────────────────────────────
#> "Month" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> Month5 67.277 [64.959, 69.596] (1.172)
#> Month6 79.196 [77.048, 81.343] (1.086)
#> Month7 82.412 [80.252, 84.573] (1.092)
#> Month8 83.450 [81.201, 85.698] (1.137)
#> Month9 77.465 [75.309, 79.621] (1.090)
#> ────────────────────────────────────────
#>
#> Consecutive (Sequential) Comparisons of "Month":
#> ──────────────────────────────────────────────────────────────────────────────
#> Contrast Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────────
#> Month6 - Month5 11.918 (1.592) 134 7.486 <.001 *** 2.006 [ 1.327, 2.684]
#> Month7 - Month6 3.217 (1.541) 134 2.087 .155 0.541 [-0.115, 1.198]
#> Month8 - Month7 1.038 (1.573) 134 0.660 1.000 0.175 [-0.496, 0.845]
#> Month9 - Month8 -5.985 (1.571) 134 -3.808 <.001 *** -1.007 [-1.677, -0.338]
#> ──────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 5.942
#> Results are averaged over the levels of: Day.1or2
#> P-value adjustment: Bonferroni method for 4 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "Month") ------
#>
#> Joint Tests of "Month":
#> ────────────────────────────────────────────────────────────────
#> Effect "Day.1or2" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────────────
#> Month odd 4 134 13.278 <.001 *** .284 [.169, .371]
#> Month even 4 134 18.140 <.001 *** .351 [.236, .437]
#> Solar.R odd 1 134 17.359 <.001 *** .115 [.043, .204]
#> Solar.R even 1 134 17.359 <.001 *** .115 [.043, .204]
#> Wind odd 1 134 24.925 <.001 *** .157 [.074, .251]
#> Wind even 1 134 24.925 <.001 *** .157 [.074, .251]
#> ────────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "Month":
#> ──────────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ──────────────────────────────────────────────────────────────
#> odd: "Month" 1875.512 4 468.878 13.278 <.001 ***
#> even: "Month" 2562.180 4 640.545 18.140 <.001 ***
#> Residuals 4731.762 134 35.312
#> ──────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "Month":
#> ───────────────────────────────────────────────────
#> "Month" "Day.1or2" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────────────────
#> Month5 odd 67.644 [64.342, 70.946] (1.669)
#> Month6 odd 79.321 [76.275, 82.367] (1.540)
#> Month7 odd 81.991 [79.019, 84.962] (1.503)
#> Month8 odd 82.494 [79.416, 85.573] (1.557)
#> Month9 odd 77.268 [74.229, 80.307] (1.536)
#> Month5 even 66.910 [63.732, 70.089] (1.607)
#> Month6 even 79.070 [76.032, 82.109] (1.536)
#> Month7 even 82.834 [79.746, 85.922] (1.561)
#> Month8 even 84.405 [81.135, 87.676] (1.654)
#> Month9 even 77.663 [74.613, 80.713] (1.542)
#> ───────────────────────────────────────────────────
#>
#> Polynomial Contrasts of "Month":
#> ────────────────────────────────────────────────────────────
#> Contrast "Day.1or2" Estimate S.E. df t p
#> ────────────────────────────────────────────────────────────
#> linear odd 22.420 ( 5.061) 134 4.430 <.001 ***
#> quadratic odd -35.973 ( 5.946) 134 -6.050 <.001 ***
#> cubic odd 3.277 ( 4.925) 134 0.665 .507
#> quartic odd -10.405 (12.661) 134 -0.822 .413
#> linear even 26.840 ( 5.008) 134 5.359 <.001 ***
#> quadratic even -39.997 ( 5.922) 134 -6.754 <.001 ***
#> cubic even 0.082 ( 5.034) 134 0.016 .987
#> quartic even -12.327 (13.192) 134 -0.934 .352
#> ────────────────────────────────────────────────────────────
#>
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
# }
between.3
#> A B C SCORE
#> 1 1 1 1 3
#> 2 1 1 1 6
#> 3 1 1 1 4
#> 4 1 1 1 3
#> 5 1 1 2 5
#> 6 1 1 2 7
#> 7 1 1 2 5
#> 8 1 1 2 2
#> 9 1 2 1 4
#> 10 1 2 1 6
#> 11 1 2 1 4
#> 12 1 2 1 2
#> 13 1 2 2 4
#> 14 1 2 2 5
#> 15 1 2 2 3
#> 16 1 2 2 3
#> 17 2 1 1 8
#> 18 2 1 1 9
#> 19 2 1 1 8
#> 20 2 1 1 7
#> 21 2 1 2 5
#> 22 2 1 2 6
#> 23 2 1 2 7
#> 24 2 1 2 6
#> 25 2 2 1 9
#> 26 2 2 1 8
#> 27 2 2 1 8
#> 28 2 2 1 7
#> 29 2 2 2 12
#> 30 2 2 2 13
#> 31 2 2 2 12
#> 32 2 2 2 11
MANOVA(between.3, dv="SCORE", between=c("A", "B", "C")) %>%
EMMEANS("A", by="B") %>%
EMMEANS(c("A", "B"), by="C") %>%
EMMEANS("A", by=c("B", "C"))
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────────
#> "A" "B" "C" Mean S.D. n
#> ─────────────────────────────
#> A1 B1 C1 4.000 (1.414) 4
#> A1 B1 C2 4.750 (2.062) 4
#> A1 B2 C1 4.000 (1.633) 4
#> A1 B2 C2 3.750 (0.957) 4
#> A2 B1 C1 8.000 (0.816) 4
#> A2 B1 C2 6.000 (0.816) 4
#> A2 B2 C1 8.000 (0.816) 4
#> A2 B2 C2 12.000 (0.816) 4
#> ─────────────────────────────
#> Total sample size: N = 32
#>
#> ANOVA Table:
#> Dependent variable(s): SCORE
#> Between-subjects factor(s): A, B, C
#> Within-subjects factor(s): –
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 153.125 1.563 1 24 98.000 <.001 *** .803 [.670, .870] .803
#> B 12.500 1.563 1 24 8.000 .009 ** .250 [.042, .466] .250
#> C 3.125 1.563 1 24 2.000 .170 .077 [.000, .283] .077
#> A * B 24.500 1.563 1 24 15.680 <.001 *** .395 [.147, .585] .395
#> A * C 1.125 1.563 1 24 0.720 .405 .029 [.000, .206] .029
#> B * C 12.500 1.563 1 24 8.000 .009 ** .250 [.042, .466] .250
#> A * B * C 24.500 1.563 1 24 15.680 <.001 *** .395 [.147, .585] .395
#> ──────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ───────────────────────────────────────
#> Levene’s F df1 df2 p
#> ───────────────────────────────────────
#> DV: SCORE 0.668 7 24 .697
#> ───────────────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A B1 1 24 17.640 <.001 *** .424 [.173, .607]
#> A B2 1 24 96.040 <.001 *** .800 [.665, .868]
#> C B1 1 24 1.000 .327 .040 [.000, .226]
#> C B2 1 24 9.000 .006 ** .273 [.055, .486]
#> A * C B1 1 24 4.840 .038 * .168 [.006, .388]
#> A * C B2 1 24 11.560 .002 ** .325 [.090, .530]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────
#> B1: "A" 27.563 1 27.563 17.640 <.001 ***
#> B2: "A" 150.062 1 150.062 96.040 <.001 ***
#> Residuals 37.500 24 1.563
#> ─────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────
#> A1 B1 4.375 [3.463, 5.287] (0.442)
#> A2 B1 7.000 [6.088, 7.912] (0.442)
#> A1 B2 3.875 [2.963, 4.787] (0.442)
#> A2 B2 10.000 [9.088, 10.912] (0.442)
#> ───────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ─────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ─────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 2.625 (0.625) 24 4.200 <.001 *** 2.100 [1.068, 3.132]
#> A2 - A1 B2 6.125 (0.625) 24 9.800 <.001 *** 4.900 [3.868, 5.932]
#> ─────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.250
#> Results are averaged over the levels of: C
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A" & "B") ------
#>
#> Joint Tests of "A" & "B":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A C1 1 24 40.960 <.001 *** .631 [.414, .754]
#> A C2 1 24 57.760 <.001 *** .706 [.521, .806]
#> B C1 1 24 0.000 1.000 .000 [.000, .000]
#> B C2 1 24 16.000 <.001 *** .400 [.151, .589]
#> A * B C1 1 24 0.000 1.000 .000 [.000, .000]
#> A * B C2 1 24 31.360 <.001 *** .566 [.331, .710]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A" & "B":
#> ─────────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ─────────────────────────────────────────────────────────────
#> C1: "A" & "B" 0.000 1 0.000 0.000 1.000
#> C2: "A" & "B" 49.000 1 49.000 31.360 <.001 ***
#> Residuals 37.500 24 1.563
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A" & "B":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 2.710, 5.290] (0.625)
#> A2 B1 8.000 [ 6.710, 9.290] (0.625)
#> A1 B2 4.000 [ 2.710, 5.290] (0.625)
#> A2 B2 8.000 [ 6.710, 9.290] (0.625)
#> A1 B1 4.750 [ 3.460, 6.040] (0.625)
#> A2 B1 6.000 [ 4.710, 7.290] (0.625)
#> A1 B2 3.750 [ 2.460, 5.040] (0.625)
#> A2 B2 12.000 [10.710, 13.290] (0.625)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A" & "B":
#> ───────────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ───────────────────────────────────────────────────────────────────────────────
#> A2 B1 - A1 B1 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.167, 5.233]
#> A1 B2 - A1 B1 C1 0.000 (0.884) 24 0.000 1.000 0.000 [-2.033, 2.033]
#> A1 B2 - A2 B1 C1 -4.000 (0.884) 24 -4.525 <.001 *** -3.200 [-5.233, -1.167]
#> A2 B2 - A1 B1 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.167, 5.233]
#> A2 B2 - A2 B1 C1 -0.000 (0.884) 24 -0.000 1.000 -0.000 [-2.033, 2.033]
#> A2 B2 - A1 B2 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.167, 5.233]
#> A2 B1 - A1 B1 C2 1.250 (0.884) 24 1.414 1.000 1.000 [-1.033, 3.033]
#> A1 B2 - A1 B1 C2 -1.000 (0.884) 24 -1.131 1.000 -0.800 [-2.833, 1.233]
#> A1 B2 - A2 B1 C2 -2.250 (0.884) 24 -2.546 .107 -1.800 [-3.833, 0.233]
#> A2 B2 - A1 B1 C2 7.250 (0.884) 24 8.202 <.001 *** 5.800 [ 3.767, 7.833]
#> A2 B2 - A2 B1 C2 6.000 (0.884) 24 6.788 <.001 *** 4.800 [ 2.767, 6.833]
#> A2 B2 - A1 B2 C2 8.250 (0.884) 24 9.334 <.001 *** 6.600 [ 4.567, 8.633]
#> ───────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.250
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────────
#> Effect "B" "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────────
#> A B1 C1 1 24 20.480 <.001 *** .460 [.210, .634]
#> A B2 C1 1 24 20.480 <.001 *** .460 [.210, .634]
#> A B1 C2 1 24 2.000 .170 .077 [.000, .283]
#> A B2 C2 1 24 87.120 <.001 *** .784 [.639, .858]
#> ────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "A":
#> ────────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ────────────────────────────────────────────────────────────
#> B1 & C1: "A" 32.000 1 32.000 20.480 <.001 ***
#> B2 & C1: "A" 32.000 1 32.000 20.480 <.001 ***
#> B1 & C2: "A" 3.125 1 3.125 2.000 .170
#> B2 & C2: "A" 136.125 1 136.125 87.120 <.001 ***
#> Residuals 37.500 24 1.563
#> ────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────────
#> "A" "B" "C" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────────
#> A1 B1 C1 4.000 [ 2.710, 5.290] (0.625)
#> A2 B1 C1 8.000 [ 6.710, 9.290] (0.625)
#> A1 B2 C1 4.000 [ 2.710, 5.290] (0.625)
#> A2 B2 C1 8.000 [ 6.710, 9.290] (0.625)
#> A1 B1 C2 4.750 [ 3.460, 6.040] (0.625)
#> A2 B1 C2 6.000 [ 4.710, 7.290] (0.625)
#> A1 B2 C2 3.750 [ 2.460, 5.040] (0.625)
#> A2 B2 C2 12.000 [10.710, 13.290] (0.625)
#> ────────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ─────────────────────────────────────────────────────────────────────────────
#> Contrast "B" "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ─────────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.741, 4.659]
#> A2 - A1 B2 C1 4.000 (0.884) 24 4.525 <.001 *** 3.200 [ 1.741, 4.659]
#> A2 - A1 B1 C2 1.250 (0.884) 24 1.414 .170 1.000 [-0.459, 2.459]
#> A2 - A1 B2 C2 8.250 (0.884) 24 9.334 <.001 *** 6.600 [ 5.141, 8.059]
#> ─────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.250
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
## Just to name a few...
## You may test other combinations...
#### Within-Subjects Design ####
within.1
#> ID A1 A2 A3 A4
#> 1 S1 3 4 8 9
#> 2 S2 6 6 9 8
#> 3 S3 4 4 8 8
#> 4 S4 3 2 7 7
#> 5 S5 5 4 5 12
#> 6 S6 7 5 6 13
#> 7 S7 5 3 7 12
#> 8 S8 2 3 6 11
MANOVA(within.1, dvs="A1:A4", dvs.pattern="A(.)",
within="A") %>%
EMMEANS("A")
#>
#> Note:
#> dvs="A1:A4" is matched to variables:
#> A1, A2, A3, A4
#>
#> ====== ANOVA (Within-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────
#> "A" Mean S.D. n
#> ─────────────────────
#> A1 4.375 (1.685) 8
#> A2 3.875 (1.246) 8
#> A3 7.000 (1.309) 8
#> A4 10.000 (2.268) 8
#> ─────────────────────
#> Total sample size: N = 8
#>
#> ANOVA Table:
#> Dependent variable(s): A1, A2, A3, A4
#> Between-subjects factor(s): –
#> Within-subjects factor(s): A
#> Covariate(s): –
#> ─────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ─────────────────────────────────────────────────────────────────
#> A 63.375 2.518 3 21 25.170 <.001 *** .782 [.609, .858] .707
#> ─────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> No between-subjects factors. No need to do the Levene’s test.
#>
#> Mauchly’s Test of Sphericity:
#> ────────────────────────
#> Mauchly's W p
#> ────────────────────────
#> A 0.1899 .095 .
#> ────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────
#> Effect df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────
#> A 3 7 47.960 <.001 *** .954 [.848, .977]
#> ────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ───────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────
#> Mean: "A" 0.954 3.000 5.000 34.257 <.001 ***
#> ───────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────
#> "A" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────
#> A1 4.375 [2.966, 5.784] (0.596)
#> A2 3.875 [2.833, 4.917] (0.441)
#> A3 7.000 [5.905, 8.095] (0.463)
#> A4 10.000 [8.104, 11.896] (0.802)
#> ───────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────
#> Contrast Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────
#> A2 - A1 -0.500 (0.423) 7 -1.183 1.000 -0.223 [-0.907, 0.462]
#> A3 - A1 2.625 (0.754) 7 3.479 .062 . 1.170 [-0.053, 2.392]
#> A3 - A2 3.125 (0.515) 7 6.063 .003 ** 1.393 [ 0.558, 2.228]
#> A4 - A1 5.625 (0.778) 7 7.232 .001 ** 2.507 [ 1.247, 3.767]
#> A4 - A2 6.125 (0.875) 7 7.000 .001 ** 2.729 [ 1.312, 4.147]
#> A4 - A3 3.000 (1.180) 7 2.542 .231 1.337 [-0.575, 3.249]
#> ──────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 2.244
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
within.2
#> ID A1B1 A1B2 A1B3 A2B1 A2B2 A2B3
#> 1 S1 3 4 5 4 8 12
#> 2 S2 6 6 7 5 9 13
#> 3 S3 4 4 5 3 8 12
#> 4 S4 3 2 2 3 7 11
MANOVA(within.2, dvs="A1B1:A2B3", dvs.pattern="A(.)B(.)",
within=c("A", "B")) %>%
EMMEANS("A", by="B") %>%
EMMEANS("B", by="A") # singular error matrix
#>
#> Note:
#> dvs="A1B1:A2B3" is matched to variables:
#> A1B1, A1B2, A1B3, A2B1, A2B2, A2B3
#>
#> ====== ANOVA (Within-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────
#> "A" "B" Mean S.D. n
#> ─────────────────────────
#> A1 B1 4.000 (1.414) 4
#> A1 B2 4.000 (1.633) 4
#> A1 B3 4.750 (2.062) 4
#> A2 B1 3.750 (0.957) 4
#> A2 B2 8.000 (0.816) 4
#> A2 B3 12.000 (0.816) 4
#> ─────────────────────────
#> Total sample size: N = 4
#>
#> ANOVA Table:
#> Dependent variable(s): A1B1, A1B2, A1B3, A2B1, A2B2, A2B3
#> Between-subjects factor(s): –
#> Within-subjects factor(s): A, B
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────
#> A 80.667 1.111 1 3 72.600 .003 ** .960 [.699, .985] .707
#> B 40.542 0.264 2 6 153.632 <.001 *** .981 [.930, .991] .708
#> A * B 28.292 0.236 2 6 119.824 <.001 *** .976 [.911, .988] .628
#> ──────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> No between-subjects factors. No need to do the Levene’s test.
#>
#> Mauchly’s Test of Sphericity:
#> ────────────────────────────
#> Mauchly's W p
#> ────────────────────────────
#> B 0.0665 .066 .
#> A * B 0.2491 .249
#> ────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────
#> A B1 1 3 0.273 .638 .083 [.000, .597]
#> A B2 1 3 96.000 .002 ** .970 [.763, .988]
#> A B3 1 3 132.789 .001 ** .978 [.823, .992]
#> ─────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.083 1.000 3.000 0.273 .638
#> B2: "A" 0.970 1.000 3.000 96.000 .002 **
#> B3: "A" 0.978 1.000 3.000 132.789 .001 **
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 1.750, 6.250] (0.707)
#> A2 B1 3.750 [ 2.227, 5.273] (0.479)
#> A1 B2 4.000 [ 1.402, 6.598] (0.816)
#> A2 B2 8.000 [ 6.701, 9.299] (0.408)
#> A1 B3 4.750 [ 1.470, 8.030] (1.031)
#> A2 B3 12.000 [10.701, 13.299] (0.408)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 -0.250 (0.479) 3 -0.522 .638 -0.272 [-1.930, 1.386]
#> A2 - A1 B2 4.000 (0.408) 3 9.798 .002 ** 4.353 [ 2.939, 5.767]
#> A2 - A1 B3 7.250 (0.629) 3 11.523 .001 ** 7.890 [ 5.711, 10.068]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.919
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "B") ------
#>
#> Error in solve.default(zcov, z) :
#> system is computationally singular: reciprocal condition number = 2.23113e-17
#> Error in solve.default(zcov, z) :
#> system is computationally singular: reciprocal condition number = 2.23113e-17
#> Warning: Some CIs could not be estimated due to non-finite F, df, or df_error
#> values.
#> Joint Tests of "B":
#> ───────────────────────────────────────────────────────────────
#> Effect "A" df1 df2 F p η²p [90% CI of η²p]
#> ───────────────────────────────────────────────────────────────
#> B A1 2 3 13.500 .032 * .900 [.219, .962]
#> B A2 2 NA [ NA, NA]
#> (confounded) A2 2 NA [ NA, NA]
#> ───────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Estimated Marginal Means of "B":
#> ────────────────────────────────────────
#> "B" "A" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> B1 A1 4.000 [ 1.750, 6.250] (0.707)
#> B2 A1 4.000 [ 1.402, 6.598] (0.816)
#> B3 A1 4.750 [ 1.470, 8.030] (1.031)
#> B1 A2 3.750 [ 2.227, 5.273] (0.479)
#> B2 A2 8.000 [ 6.701, 9.299] (0.408)
#> B3 A2 12.000 [10.701, 13.299] (0.408)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "B":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "A" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> B2 - B1 A1 0.000 (0.408) 3 0.000 1.000 0.000 [-2.158, 2.158]
#> B3 - B1 A1 0.750 (0.629) 3 1.192 .957 0.816 [-2.509, 4.141]
#> B3 - B2 A1 0.750 (0.250) 3 3.000 .173 0.816 [-0.505, 2.137]
#> B2 - B1 A2 4.250 (0.250) 3 17.000 .001 ** 4.625 [ 3.304, 5.946]
#> B3 - B1 A2 8.250 (0.250) 3 33.000 <.001 *** 8.978 [ 7.656, 10.299]
#> B3 - B2 A2 4.000 (0.000) 3 Inf <.001 *** 4.353 [ 4.353, 4.353]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.919
#> P-value adjustment: Bonferroni method for 3 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
# :::::::::::::::::::::::::::::::::::::::
# This would produce a WARNING because of
# the linear dependence of A2B2 and A2B3.
# See: Corr(within.2[c("A2B2", "A2B3")])
within.3
#> ID A1B1C1 A1B1C2 A1B2C1 A1B2C2 A2B1C1 A2B1C2 A2B2C1 A2B2C2
#> 1 S1 3 5 4 4 8 5 9 12
#> 2 S2 6 7 6 5 9 6 8 13
#> 3 S3 4 5 4 3 8 7 8 12
#> 4 S4 3 2 2 3 7 6 7 11
MANOVA(within.3, dvs="A1B1C1:A2B2C2", dvs.pattern="A(.)B(.)C(.)",
within=c("A", "B", "C")) %>%
EMMEANS("A", by="B") %>%
EMMEANS(c("A", "B"), by="C") %>%
EMMEANS("A", by=c("B", "C"))
#>
#> Note:
#> dvs="A1B1C1:A2B2C2" is matched to variables:
#> A1B1C1, A1B1C2, A1B2C1, A1B2C2, A2B1C1, A2B1C2, A2B2C1, A2B2C2
#>
#> ====== ANOVA (Within-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────────
#> "A" "B" "C" Mean S.D. n
#> ─────────────────────────────
#> A1 B1 C1 4.000 (1.414) 4
#> A1 B1 C2 4.750 (2.062) 4
#> A1 B2 C1 4.000 (1.633) 4
#> A1 B2 C2 3.750 (0.957) 4
#> A2 B1 C1 8.000 (0.816) 4
#> A2 B1 C2 6.000 (0.816) 4
#> A2 B2 C1 8.000 (0.816) 4
#> A2 B2 C2 12.000 (0.816) 4
#> ─────────────────────────────
#> Total sample size: N = 4
#>
#> ANOVA Table:
#> Dependent variable(s): A1B1C1, A1B1C2, A1B2C1, A1B2C2, A2B1C1, A2B1C2, A2B2C1, A2B2C2
#> Between-subjects factor(s): –
#> Within-subjects factor(s): A, B, C
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 153.125 1.875 1 3 81.667 .003 ** .965 [.727, .986] .803
#> B 12.500 0.583 1 3 21.429 .019 * .877 [.279, .954] .250
#> C 3.125 0.042 1 3 75.000 .003 ** .962 [.707, .985] .077
#> A * B 24.500 0.250 1 3 98.000 .002 ** .970 [.768, .989] .395
#> A * C 1.125 0.708 1 3 1.588 .297 .346 [.000, .751] .029
#> B * C 12.500 0.417 1 3 30.000 .012 * .909 [.411, .965] .250
#> A * B * C 24.500 1.083 1 3 22.615 .018 * .883 [.300, .956] .395
#> ──────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> No between-subjects factors. No need to do the Levene’s test.
#>
#> Mauchly’s Test of Sphericity:
#> The repeated measures have only two levels. The assumption of sphericity is always met.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────
#> C B1 1 3 6.818 .080 . .694 [.000, .886]
#> C B2 1 3 61.364 .004 ** .953 [.653, .982]
#> A B1 1 3 17.640 .025 * .855 [.202, .945]
#> A B2 1 3 266.778 <.001 *** .989 [.908, .996]
#> C * A B1 1 3 6.153 .089 . .672 [.000, .878]
#> C * A B2 1 3 32.111 .011 * .915 [.436, .968]
#> ─────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.855 1.000 3.000 17.640 .025 *
#> B2: "A" 0.989 1.000 3.000 266.778 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────
#> A1 B1 4.375 [1.746, 7.004] (0.826)
#> A2 B1 7.000 [6.081, 7.919] (0.289)
#> A1 B2 3.875 [1.886, 5.864] (0.625)
#> A2 B2 10.000 [8.875, 11.125] (0.354)
#> ───────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 2.625 (0.625) 3 4.200 .025 * 2.205 [0.534, 3.877]
#> A2 - A1 B2 6.125 (0.375) 3 16.333 <.001 *** 5.146 [4.143, 6.149]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.190
#> Results are averaged over the levels of: C
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A" & "B") ------
#>
#> Error in solve.default(zcov, z) :
#> system is computationally singular: reciprocal condition number = 1.67575e-17
#> Error in solve.default(zcov, z) :
#> system is computationally singular: reciprocal condition number = 1.38778e-17
#> Joint Tests of "A" & "B":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> B C1 1 3 0.000 1.000 .000 [.000, .000]
#> B C2 1 3 50.000 .006 ** .943 [.592, .978]
#> A C1 1 3 54.857 .005 ** .948 [.620, .980]
#> A C2 1 3 63.706 .004 ** .955 [.664, .983]
#> B * A C1 1 3 0.000 1.000 .000 [.000, .000]
#> B * A C2 1 3 42.000 .007 ** .933 [.534, .975]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A" & "B":
#> ───────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────────
#> C1: "A" & "B" 0.000 1.000 3.000 0.000 1.000
#> C2: "A" & "B" 0.933 1.000 3.000 42.000 .007 **
#> ───────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A" & "B":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 1.750, 6.250] (0.707)
#> A2 B1 8.000 [ 6.701, 9.299] (0.408)
#> A1 B2 4.000 [ 1.402, 6.598] (0.816)
#> A2 B2 8.000 [ 6.701, 9.299] (0.408)
#> A1 B1 4.750 [ 1.470, 8.030] (1.031)
#> A2 B1 6.000 [ 4.701, 7.299] (0.408)
#> A1 B2 3.750 [ 2.227, 5.273] (0.479)
#> A2 B2 12.000 [10.701, 13.299] (0.408)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A" & "B":
#> ───────────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ───────────────────────────────────────────────────────────────────────────────
#> A2 B1 - A1 B1 C1 4.000 (0.408) 3 9.798 .014 * 3.361 [ 1.223, 5.498]
#> A1 B2 - A1 B1 C1 0.000 (0.408) 3 0.000 1.000 0.000 [-2.137, 2.137]
#> A1 B2 - A2 B1 C1 -4.000 (0.408) 3 -9.798 .014 * -3.361 [-5.498, -1.223]
#> A2 B2 - A1 B1 C1 4.000 (0.816) 3 4.899 .098 . 3.361 [-0.914, 7.635]
#> A2 B2 - A2 B1 C1 0.000 (0.408) 3 0.000 1.000 0.000 [-2.137, 2.137]
#> A2 B2 - A1 B2 C1 4.000 (0.707) 3 5.657 .066 . 3.361 [-0.341, 7.063]
#> A2 B1 - A1 B1 C2 1.250 (1.109) 3 1.127 1.000 1.050 [-4.754, 6.855]
#> A1 B2 - A1 B1 C2 -1.000 (0.707) 3 -1.414 1.000 -0.840 [-4.542, 2.862]
#> A1 B2 - A2 B1 C2 -2.250 (0.750) 3 -3.000 .346 -1.890 [-5.817, 2.036]
#> A2 B2 - A1 B1 C2 7.250 (0.629) 3 11.523 .008 ** 6.091 [ 2.797, 9.385]
#> A2 B2 - A2 B1 C2 6.000 (0.577) 3 10.392 .011 * 5.041 [ 2.018, 8.064]
#> A2 B2 - A1 B2 C2 8.250 (0.250) 3 33.000 <.001 *** 6.931 [ 5.623, 8.240]
#> ───────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.190
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ──────────────────────────────────────────────────────────────
#> Effect "B" "C" df1 df2 F p η²p [90% CI of η²p]
#> ──────────────────────────────────────────────────────────────
#> A B1 C1 1 3 96.000 .002 ** .970 [.763, .988]
#> A B2 C1 1 3 32.000 .011 * .914 [.435, .967]
#> A B1 C2 1 3 1.271 .342 .298 [.000, .729]
#> A B2 C2 1 3 1089.000 <.001 *** .997 [.977, .999]
#> ──────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ───────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────────
#> B1 & C1: "A" 0.970 1.000 3.000 96.000 .002 **
#> B2 & C1: "A" 0.914 1.000 3.000 32.000 .011 *
#> B1 & C2: "A" 0.298 1.000 3.000 1.271 .342
#> B2 & C2: "A" 0.997 1.000 3.000 1089.000 <.001 ***
#> ───────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────────
#> "A" "B" "C" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────────
#> A1 B1 C1 4.000 [ 1.750, 6.250] (0.707)
#> A2 B1 C1 8.000 [ 6.701, 9.299] (0.408)
#> A1 B2 C1 4.000 [ 1.402, 6.598] (0.816)
#> A2 B2 C1 8.000 [ 6.701, 9.299] (0.408)
#> A1 B1 C2 4.750 [ 1.470, 8.030] (1.031)
#> A2 B1 C2 6.000 [ 4.701, 7.299] (0.408)
#> A1 B2 C2 3.750 [ 2.227, 5.273] (0.479)
#> A2 B2 C2 12.000 [10.701, 13.299] (0.408)
#> ────────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────────
#> Contrast "B" "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 C1 4.000 (0.408) 3 9.798 .002 ** 3.361 [ 2.269, 4.452]
#> A2 - A1 B2 C1 4.000 (0.707) 3 5.657 .011 * 3.361 [ 1.470, 5.251]
#> A2 - A1 B1 C2 1.250 (1.109) 3 1.127 .342 1.050 [-1.914, 4.015]
#> A2 - A1 B2 C2 8.250 (0.250) 3 33.000 <.001 *** 6.931 [ 6.263, 7.600]
#> ──────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 1.190
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
## Just to name a few...
## You may test other combinations...
#### Mixed Design ####
mixed.2_1b1w
#> A B1 B2 B3
#> 1 1 3 4 5
#> 2 1 6 6 7
#> 3 1 4 4 5
#> 4 1 3 2 2
#> 5 2 4 8 12
#> 6 2 5 9 13
#> 7 2 3 8 12
#> 8 2 3 7 11
MANOVA(mixed.2_1b1w, dvs="B1:B3", dvs.pattern="B(.)",
between="A", within="B", sph.correction="GG") %>%
EMMEANS("A", by="B") %>%
EMMEANS("B", by="A")
#>
#> Note:
#> dvs="B1:B3" is matched to variables:
#> B1, B2, B3
#>
#> ====== ANOVA (Mixed Design) ======
#>
#> Descriptives:
#> ─────────────────────────
#> "A" "B" Mean S.D. n
#> ─────────────────────────
#> A1 B1 4.000 (1.414) 4
#> A1 B2 4.000 (1.633) 4
#> A1 B3 4.750 (2.062) 4
#> A2 B1 3.750 (0.957) 4
#> A2 B2 8.000 (0.816) 4
#> A2 B3 12.000 (0.816) 4
#> ─────────────────────────
#> Total sample size: N = 8
#>
#> ANOVA Table:
#> Dependent variable(s): B1, B2, B3
#> Between-subjects factor(s): A
#> Within-subjects factor(s): B
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 80.667 5.083 1.000 6.000 15.869 .007 ** .726 [.248, .871] .707
#> B 74.702 0.461 1.085 6.513 162.167 <.001 *** .964 [.880, .983] .708
#> A * B 52.130 0.461 1.085 6.513 113.167 <.001 *** .950 [.833, .976] .628
#> ──────────────────────────────────────────────────────────────────────────
#> Sphericity correction method: GG (Greenhouse-Geisser)
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ────────────────────────────────────
#> Levene’s F df1 df2 p
#> ────────────────────────────────────
#> DV: B1 0.300 1 6 .604
#> DV: B2 0.600 1 6 .468
#> DV: B3 1.485 1 6 .269
#> ────────────────────────────────────
#>
#> Mauchly’s Test of Sphericity:
#> ────────────────────────────
#> Mauchly's W p
#> ────────────────────────────
#> B 0.1574 .010 **
#> A * B 0.1574 .010 **
#> ────────────────────────────
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A B1 1 6 0.086 .780 .014 [.000, .339]
#> A B2 1 6 19.200 .005 ** .762 [.314, .888]
#> A B3 1 6 42.763 <.001 *** .877 [.593, .942]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.014 1.000 6.000 0.086 .780
#> B2: "A" 0.762 1.000 6.000 19.200 .005 **
#> B3: "A" 0.877 1.000 6.000 42.763 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 2.523, 5.477] (0.604)
#> A2 B1 3.750 [ 2.273, 5.227] (0.604)
#> A1 B2 4.000 [ 2.421, 5.579] (0.645)
#> A2 B2 8.000 [ 6.421, 9.579] (0.645)
#> A1 B3 4.750 [ 2.832, 6.668] (0.784)
#> A2 B3 12.000 [10.082, 13.918] (0.784)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 -0.250 (0.854) 6 -0.293 .780 -0.382 [-3.574, 2.810]
#> A2 - A1 B2 4.000 (0.913) 6 4.382 .005 ** 6.110 [ 2.698, 9.522]
#> A2 - A1 B3 7.250 (1.109) 6 6.539 <.001 *** 11.075 [ 6.931, 15.218]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.655
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "B") ------
#>
#> Joint Tests of "B":
#> ─────────────────────────────────────────────────────────
#> Effect "A" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────
#> B A1 2 6 17.471 .003 ** .853 [.492, .930]
#> B A2 2 6 265.941 <.001 *** .989 [.959, .995]
#> ─────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "B":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> A1: "B" 0.853 2.000 5.000 14.559 .008 **
#> A2: "B" 0.989 2.000 5.000 221.618 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "B":
#> ────────────────────────────────────────
#> "B" "A" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> B1 A1 4.000 [ 2.523, 5.477] (0.604)
#> B2 A1 4.000 [ 2.421, 5.579] (0.645)
#> B3 A1 4.750 [ 2.832, 6.668] (0.784)
#> B1 A2 3.750 [ 2.273, 5.227] (0.604)
#> B2 A2 8.000 [ 6.421, 9.579] (0.645)
#> B3 A2 12.000 [10.082, 13.918] (0.784)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "B":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "A" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> B2 - B1 A1 -0.000 (0.339) 6 -0.000 1.000 -0.000 [-1.700, 1.700]
#> B3 - B1 A1 0.750 (0.479) 6 1.567 .505 1.146 [-1.258, 3.550]
#> B3 - B2 A1 0.750 (0.177) 6 4.243 .016 * 1.146 [ 0.258, 2.033]
#> B2 - B1 A2 4.250 (0.339) 6 12.555 <.001 *** 6.492 [ 4.792, 8.192]
#> B3 - B1 A2 8.250 (0.479) 6 17.234 <.001 *** 12.602 [10.198, 15.006]
#> B3 - B2 A2 4.000 (0.177) 6 22.627 <.001 *** 6.110 [ 5.222, 6.998]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.655
#> P-value adjustment: Bonferroni method for 3 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
mixed.3_1b2w
#> A B1C1 B1C2 B2C1 B2C2
#> 1 1 3 5 4 4
#> 2 1 6 7 6 5
#> 3 1 4 5 4 3
#> 4 1 3 2 2 3
#> 5 2 8 5 9 12
#> 6 2 9 6 8 13
#> 7 2 8 7 8 12
#> 8 2 7 6 7 11
MANOVA(mixed.3_1b2w, dvs="B1C1:B2C2", dvs.pattern="B(.)C(.)",
between="A", within=c("B", "C")) %>%
EMMEANS("A", by="B") %>%
EMMEANS(c("A", "B"), by="C") %>%
EMMEANS("A", by=c("B", "C"))
#>
#> Note:
#> dvs="B1C1:B2C2" is matched to variables:
#> B1C1, B1C2, B2C1, B2C2
#>
#> ====== ANOVA (Mixed Design) ======
#>
#> Descriptives:
#> ─────────────────────────────
#> "A" "B" "C" Mean S.D. n
#> ─────────────────────────────
#> A1 B1 C1 4.000 (1.414) 4
#> A1 B1 C2 4.750 (2.062) 4
#> A1 B2 C1 4.000 (1.633) 4
#> A1 B2 C2 3.750 (0.957) 4
#> A2 B1 C1 8.000 (0.816) 4
#> A2 B1 C2 6.000 (0.816) 4
#> A2 B2 C1 8.000 (0.816) 4
#> A2 B2 C2 12.000 (0.816) 4
#> ─────────────────────────────
#> Total sample size: N = 8
#>
#> ANOVA Table:
#> Dependent variable(s): B1C1, B1C2, B2C1, B2C2
#> Between-subjects factor(s): A
#> Within-subjects factor(s): B, C
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 153.125 4.708 1 6 32.522 .001 ** .844 [.503, .926] .803
#> B 12.500 0.417 1 6 30.000 .002 ** .833 [.475, .921] .250
#> A * B 24.500 0.417 1 6 58.800 <.001 *** .907 [.684, .956] .395
#> C 3.125 0.375 1 6 8.333 .028 * .581 [.064, .801] .077
#> A * C 1.125 0.375 1 6 3.000 .134 .333 [.000, .671] .029
#> B * C 12.500 0.750 1 6 16.667 .006 ** .735 [.264, .875] .250
#> A * B * C 24.500 0.750 1 6 32.667 .001 ** .845 [.505, .927] .395
#> ──────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ──────────────────────────────────────
#> Levene’s F df1 df2 p
#> ──────────────────────────────────────
#> DV: B1C1 1.000 1 6 .356
#> DV: B1C2 1.485 1 6 .269
#> DV: B2C1 0.600 1 6 .468
#> DV: B2C2 0.500 1 6 .506
#> ──────────────────────────────────────
#>
#> Mauchly’s Test of Sphericity:
#> The repeated measures have only two levels. The assumption of sphericity is always met.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A B1 1 6 9.000 .024 * .600 [.082, .810]
#> A B2 1 6 72.758 <.001 *** .924 [.736, .964]
#> C B1 1 6 2.143 .194 .263 [.000, .629]
#> C B2 1 6 35.526 <.001 *** .856 [.533, .932]
#> A * C B1 1 6 10.371 .018 * .633 [.117, .827]
#> A * C B2 1 6 45.632 <.001 *** .884 [.613, .945]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.600 1.000 6.000 9.000 .024 *
#> B2: "A" 0.924 1.000 6.000 72.758 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────
#> A1 B1 4.375 [2.861, 5.889] (0.619)
#> A2 B1 7.000 [5.486, 8.514] (0.619)
#> A1 B2 3.875 [2.633, 5.117] (0.508)
#> A2 B2 10.000 [8.758, 11.242] (0.508)
#> ───────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ─────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ─────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 2.625 (0.875) 6 3.000 .024 * 2.797 [0.516, 5.078]
#> A2 - A1 B2 6.125 (0.718) 6 8.530 <.001 *** 6.526 [4.654, 8.398]
#> ─────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.939
#> Results are averaged over the levels of: C
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A" & "B") ------
#>
#> Joint Tests of "A" & "B":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A C1 1 6 24.000 .003 ** .800 [.395, .906]
#> A C2 1 6 37.345 <.001 *** .862 [.550, .935]
#> B C1 1 6 0.000 1.000 .000 [.000, .000]
#> B C2 1 6 30.000 .002 ** .833 [.475, .921]
#> A * B C1 1 6 0.000 1.000 .000 [.000, .000]
#> A * B C2 1 6 58.800 <.001 *** .907 [.684, .956]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A" & "B":
#> ───────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────────
#> C1: "A" & "B" 0.000 1.000 6.000 0.000 1.000
#> C2: "A" & "B" 0.907 1.000 6.000 58.800 <.001 ***
#> ───────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A" & "B":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 2.587, 5.413] (0.577)
#> A2 B1 8.000 [ 6.587, 9.413] (0.577)
#> A1 B2 4.000 [ 2.421, 5.579] (0.645)
#> A2 B2 8.000 [ 6.421, 9.579] (0.645)
#> A1 B1 4.750 [ 2.832, 6.668] (0.784)
#> A2 B1 6.000 [ 4.082, 7.918] (0.784)
#> A1 B2 3.750 [ 2.661, 4.839] (0.445)
#> A2 B2 12.000 [10.911, 13.089] (0.445)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A" & "B":
#> ───────────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ───────────────────────────────────────────────────────────────────────────────
#> A2 B1 - A1 B1 C1 4.000 (0.816) 6 4.899 .016 * 4.262 [ 0.901, 7.622]
#> A1 B2 - A1 B1 C1 0.000 (0.408) 6 0.000 1.000 0.000 [-1.680, 1.680]
#> A1 B2 - A2 B1 C1 -4.000 (0.866) 6 -4.619 .022 * -4.262 [-7.826, -0.697]
#> A2 B2 - A1 B1 C1 4.000 (0.866) 6 4.619 .022 * 4.262 [ 0.697, 7.826]
#> A2 B2 - A2 B1 C1 0.000 (0.408) 6 0.000 1.000 0.000 [-1.680, 1.680]
#> A2 B2 - A1 B2 C1 4.000 (0.913) 6 4.382 .028 * 4.262 [ 0.505, 8.019]
#> A2 B1 - A1 B1 C2 1.250 (1.109) 6 1.127 1.000 1.332 [-3.231, 5.895]
#> A1 B2 - A1 B1 C2 -1.000 (0.645) 6 -1.549 1.000 -1.065 [-3.722, 1.591]
#> A1 B2 - A2 B1 C2 -2.250 (0.901) 6 -2.496 .281 -2.397 [-6.107, 1.313]
#> A2 B2 - A1 B1 C2 7.250 (0.901) 6 8.043 .001 ** 7.724 [ 4.014, 11.434]
#> A2 B2 - A2 B1 C2 6.000 (0.645) 6 9.295 <.001 *** 6.393 [ 3.736, 9.049]
#> A2 B2 - A1 B2 C2 8.250 (0.629) 6 13.113 <.001 *** 8.790 [ 6.200, 11.379]
#> ───────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.939
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Effect "B" "C" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────────
#> A B1 C1 1 6 24.000 .003 ** .800 [.395, .906]
#> A B2 C1 1 6 19.200 .005 ** .762 [.314, .888]
#> A B1 C2 1 6 1.271 .303 .175 [.000, .568]
#> A B2 C2 1 6 171.947 <.001 *** .966 [.879, .984]
#> ─────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ──────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ──────────────────────────────────────────────────────────────────
#> B1 & C1: "A" 0.800 1.000 6.000 24.000 .003 **
#> B2 & C1: "A" 0.762 1.000 6.000 19.200 .005 **
#> B1 & C2: "A" 0.175 1.000 6.000 1.271 .303
#> B2 & C2: "A" 0.966 1.000 6.000 171.947 <.001 ***
#> ──────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ────────────────────────────────────────────
#> "A" "B" "C" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────────
#> A1 B1 C1 4.000 [ 2.587, 5.413] (0.577)
#> A2 B1 C1 8.000 [ 6.587, 9.413] (0.577)
#> A1 B2 C1 4.000 [ 2.421, 5.579] (0.645)
#> A2 B2 C1 8.000 [ 6.421, 9.579] (0.645)
#> A1 B1 C2 4.750 [ 2.832, 6.668] (0.784)
#> A2 B1 C2 6.000 [ 4.082, 7.918] (0.784)
#> A1 B2 C2 3.750 [ 2.661, 4.839] (0.445)
#> A2 B2 C2 12.000 [10.911, 13.089] (0.445)
#> ────────────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────────
#> Contrast "B" "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 C1 4.000 (0.816) 6 4.899 .003 ** 4.262 [ 2.133, 6.390]
#> A2 - A1 B2 C1 4.000 (0.913) 6 4.382 .005 ** 4.262 [ 1.882, 6.642]
#> A2 - A1 B1 C2 1.250 (1.109) 6 1.127 .303 1.332 [-1.559, 4.222]
#> A2 - A1 B2 C2 8.250 (0.629) 6 13.113 <.001 *** 8.790 [ 7.150, 10.430]
#> ──────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.939
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
## Just to name a few...
## You may test other combinations...
mixed.3_2b1w
#> A C B1 B2
#> 1 1 1 3 4
#> 2 1 1 6 6
#> 3 1 1 4 4
#> 4 1 1 3 2
#> 5 1 2 5 4
#> 6 1 2 7 5
#> 7 1 2 5 3
#> 8 1 2 2 3
#> 9 2 1 8 9
#> 10 2 1 9 8
#> 11 2 1 8 8
#> 12 2 1 7 7
#> 13 2 2 5 12
#> 14 2 2 6 13
#> 15 2 2 7 12
#> 16 2 2 6 11
MANOVA(mixed.3_2b1w, dvs="B1:B2", dvs.pattern="B(.)",
between=c("A", "C"), within="B") %>%
EMMEANS("A", by="B") %>%
EMMEANS("A", by="C") %>%
EMMEANS(c("A", "B"), by="C") %>%
EMMEANS("B", by=c("A", "C"))
#>
#> Note:
#> dvs="B1:B2" is matched to variables:
#> B1, B2
#>
#> ====== ANOVA (Mixed Design) ======
#>
#> Descriptives:
#> ─────────────────────────────
#> "A" "C" "B" Mean S.D. n
#> ─────────────────────────────
#> A1 C1 B1 4.000 (1.414) 4
#> A1 C1 B2 4.000 (1.633) 4
#> A1 C2 B1 4.750 (2.062) 4
#> A1 C2 B2 3.750 (0.957) 4
#> A2 C1 B1 8.000 (0.816) 4
#> A2 C1 B2 8.000 (0.816) 4
#> A2 C2 B1 6.000 (0.816) 4
#> A2 C2 B2 12.000 (0.816) 4
#> ─────────────────────────────
#> Total sample size: N = 16
#>
#> ANOVA Table:
#> Dependent variable(s): B1, B2
#> Between-subjects factor(s): A, C
#> Within-subjects factor(s): B
#> Covariate(s): –
#> ──────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ──────────────────────────────────────────────────────────────────────────
#> A 153.125 2.542 1 12 60.246 <.001 *** .834 [.639, .906] .803
#> C 3.125 2.542 1 12 1.230 .289 .093 [.000, .390] .077
#> A * C 1.125 2.542 1 12 0.443 .518 .036 [.000, .305] .029
#> B 12.500 0.583 1 12 21.429 <.001 *** .641 [.308, .795] .250
#> A * B 24.500 0.583 1 12 42.000 <.001 *** .778 [.532, .874] .395
#> C * B 12.500 0.583 1 12 21.429 <.001 *** .641 [.308, .795] .250
#> A * C * B 24.500 0.583 1 12 42.000 <.001 *** .778 [.532, .874] .395
#> ──────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ────────────────────────────────────
#> Levene’s F df1 df2 p
#> ────────────────────────────────────
#> DV: B1 0.946 3 12 .449
#> DV: B2 0.423 3 12 .740
#> ────────────────────────────────────
#>
#> Mauchly’s Test of Sphericity:
#> The repeated measures have only two levels. The assumption of sphericity is always met.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ─────────────────────────────────────────────────────────
#> Effect "B" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────
#> A B1 1 12 14.538 .002 ** .548 [.190, .739]
#> A B2 1 12 122.085 <.001 *** .911 [.799, .949]
#> C B1 1 12 0.824 .382 .064 [.000, .353]
#> C B2 1 12 11.441 .005 ** .488 [.129, .702]
#> A * C B1 1 12 3.989 .069 . .249 [.000, .537]
#> A * C B2 1 12 14.695 .002 ** .550 [.193, .741]
#> ─────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> B1: "A" 0.548 1.000 12.000 14.538 .002 **
#> B2: "A" 0.911 1.000 12.000 122.085 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ───────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────
#> A1 B1 4.375 [3.314, 5.436] (0.487)
#> A2 B1 7.000 [5.939, 8.061] (0.487)
#> A1 B2 3.875 [3.021, 4.729] (0.392)
#> A2 B2 10.000 [9.146, 10.854] (0.392)
#> ───────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ──────────────────────────────────────────────────────────────────────────
#> Contrast "B" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────
#> A2 - A1 B1 2.625 (0.688) 12 3.813 .002 ** 2.717 [1.164, 4.270]
#> A2 - A1 B2 6.125 (0.554) 12 11.049 <.001 *** 6.340 [5.090, 7.590]
#> ──────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.966
#> Results are averaged over the levels of: C
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A") ------
#>
#> Joint Tests of "A":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A C1 1 12 25.180 <.001 *** .677 [.361, .816]
#> A C2 1 12 35.508 <.001 *** .747 [.477, .856]
#> B C1 1 12 0.000 1.000 .000 [.000, .000]
#> B C2 1 12 42.857 <.001 *** .781 [.538, .876]
#> A * B C1 1 12 0.000 1.000 .000 [.000, .000]
#> A * B C2 1 12 84.000 <.001 *** .875 [.724, .929]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A":
#> ─────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ─────────────────────────────────────────────────────────────
#> C1: "A" 0.677 1.000 12.000 25.180 <.001 ***
#> C2: "A" 0.747 1.000 12.000 35.508 <.001 ***
#> ─────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A":
#> ──────────────────────────────────────
#> "A" "C" Mean [95% CI of Mean] S.E.
#> ──────────────────────────────────────
#> A1 C1 4.000 [2.772, 5.228] (0.564)
#> A2 C1 8.000 [6.772, 9.228] (0.564)
#> A1 C2 4.250 [3.022, 5.478] (0.564)
#> A2 C2 9.000 [7.772, 10.228] (0.564)
#> ──────────────────────────────────────
#>
#> Pairwise Comparisons of "A":
#> ─────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ─────────────────────────────────────────────────────────────────────────
#> A2 - A1 C1 4.000 (0.797) 12 5.018 <.001 *** 4.140 [2.343, 5.938]
#> A2 - A1 C2 4.750 (0.797) 12 5.959 <.001 *** 4.917 [3.119, 6.714]
#> ─────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.966
#> Results are averaged over the levels of: B
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "A" & "B") ------
#>
#> Joint Tests of "A" & "B":
#> ────────────────────────────────────────────────────────
#> Effect "C" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────
#> A C1 1 12 25.180 <.001 *** .677 [.361, .816]
#> A C2 1 12 35.508 <.001 *** .747 [.477, .856]
#> B C1 1 12 0.000 1.000 .000 [.000, .000]
#> B C2 1 12 42.857 <.001 *** .781 [.538, .876]
#> A * B C1 1 12 0.000 1.000 .000 [.000, .000]
#> A * B C2 1 12 84.000 <.001 *** .875 [.724, .929]
#> ────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "A" & "B":
#> ───────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ───────────────────────────────────────────────────────────────────
#> C1: "A" & "B" 0.000 1.000 12.000 0.000 1.000
#> C2: "A" & "B" 0.875 1.000 12.000 84.000 <.001 ***
#> ───────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "A" & "B":
#> ────────────────────────────────────────
#> "A" "B" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> A1 B1 4.000 [ 2.500, 5.500] (0.688)
#> A2 B1 8.000 [ 6.500, 9.500] (0.688)
#> A1 B2 4.000 [ 2.792, 5.208] (0.554)
#> A2 B2 8.000 [ 6.792, 9.208] (0.554)
#> A1 B1 4.750 [ 3.250, 6.250] (0.688)
#> A2 B1 6.000 [ 4.500, 7.500] (0.688)
#> A1 B2 3.750 [ 2.542, 4.958] (0.554)
#> A2 B2 12.000 [10.792, 13.208] (0.554)
#> ────────────────────────────────────────
#>
#> Pairwise Comparisons of "A" & "B":
#> ───────────────────────────────────────────────────────────────────────────────
#> Contrast "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ───────────────────────────────────────────────────────────────────────────────
#> A2 B1 - A1 B1 C1 4.000 (0.974) 12 4.108 .009 ** 4.140 [ 0.963, 7.318]
#> A1 B2 - A1 B1 C1 -0.000 (0.540) 12 -0.000 1.000 -0.000 [-1.762, 1.762]
#> A1 B2 - A2 B1 C1 -4.000 (0.884) 12 -4.525 .004 ** -4.140 [-7.025, -1.256]
#> A2 B2 - A1 B1 C1 4.000 (0.884) 12 4.525 .004 ** 4.140 [ 1.256, 7.025]
#> A2 B2 - A2 B1 C1 0.000 (0.540) 12 0.000 1.000 0.000 [-1.762, 1.762]
#> A2 B2 - A1 B2 C1 4.000 (0.784) 12 5.102 .002 ** 4.140 [ 1.582, 6.699]
#> A2 B1 - A1 B1 C2 1.250 (0.974) 12 1.284 1.000 1.294 [-1.883, 4.471]
#> A1 B2 - A1 B1 C2 -1.000 (0.540) 12 -1.852 .533 -1.035 [-2.798, 0.727]
#> A1 B2 - A2 B1 C2 -2.250 (0.884) 12 -2.546 .154 -2.329 [-5.213, 0.555]
#> A2 B2 - A1 B1 C2 7.250 (0.884) 12 8.202 <.001 *** 7.504 [ 4.620, 10.389]
#> A2 B2 - A2 B1 C2 6.000 (0.540) 12 11.110 <.001 *** 6.211 [ 4.448, 7.973]
#> A2 B2 - A1 B2 C2 8.250 (0.784) 12 10.524 <.001 *** 8.540 [ 5.981, 11.098]
#> ───────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.966
#> P-value adjustment: Bonferroni method for 6 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "B") ------
#>
#> Joint Tests of "B":
#> ─────────────────────────────────────────────────────────────
#> Effect "A" "C" df1 df2 F p η²p [90% CI of η²p]
#> ─────────────────────────────────────────────────────────────
#> B A1 C1 1 12 0.000 1.000 .000 [.000, .000]
#> B A2 C1 1 12 0.000 1.000 .000 [.000, .000]
#> B A1 C2 1 12 3.429 .089 . .222 [.000, .515]
#> B A2 C2 1 12 123.429 <.001 *** .911 [.801, .950]
#> ─────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Multivariate Tests of "B":
#> ──────────────────────────────────────────────────────────────────
#> Pillai’s trace Hypoth. df Error df Exact F p
#> ──────────────────────────────────────────────────────────────────
#> A1 & C1: "B" 0.000 1.000 12.000 0.000 1.000
#> A2 & C1: "B" 0.000 1.000 12.000 0.000 1.000
#> A1 & C2: "B" 0.222 1.000 12.000 3.429 .089 .
#> A2 & C2: "B" 0.911 1.000 12.000 123.429 <.001 ***
#> ──────────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "B":
#> ────────────────────────────────────────────
#> "B" "A" "C" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────────
#> B1 A1 C1 4.000 [ 2.500, 5.500] (0.688)
#> B2 A1 C1 4.000 [ 2.792, 5.208] (0.554)
#> B1 A2 C1 8.000 [ 6.500, 9.500] (0.688)
#> B2 A2 C1 8.000 [ 6.792, 9.208] (0.554)
#> B1 A1 C2 4.750 [ 3.250, 6.250] (0.688)
#> B2 A1 C2 3.750 [ 2.542, 4.958] (0.554)
#> B1 A2 C2 6.000 [ 4.500, 7.500] (0.688)
#> B2 A2 C2 12.000 [10.792, 13.208] (0.554)
#> ────────────────────────────────────────────
#>
#> Pairwise Comparisons of "B":
#> ──────────────────────────────────────────────────────────────────────────────
#> Contrast "A" "C" Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────────
#> B2 - B1 A1 C1 -0.000 (0.540) 12 -0.000 1.000 -0.000 [-1.218, 1.218]
#> B2 - B1 A2 C1 0.000 (0.540) 12 0.000 1.000 0.000 [-1.218, 1.218]
#> B2 - B1 A1 C2 -1.000 (0.540) 12 -1.852 .089 . -1.035 [-2.253, 0.183]
#> B2 - B1 A2 C2 6.000 (0.540) 12 11.110 <.001 *** 6.211 [ 4.993, 7.429]
#> ──────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 0.966
#> No need to adjust p values.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
## Just to name a few...
## You may test other combinations...
#### Other Examples ####
air = airquality
air$Day.1or2 = ifelse(air$Day %% 2 == 1, 1, 2) %>%
factor(levels=1:2, labels=c("odd", "even"))
MANOVA(air, dv="Temp", between=c("Month", "Day.1or2"),
covariate=c("Solar.R", "Wind")) %>%
EMMEANS("Month", contrast="seq") %>%
EMMEANS("Month", by="Day.1or2", contrast="poly")
#> Warning: Numerical variables NOT centered on 0 (matters if variable in interaction):
#> NA, Wind
#> Warning: Missing values for 7 ID(s), which were removed before analysis:
#> 5, 6, 11, 27, 96, 97, 98
#> Below the first few rows (in wide format) of the removed cases with missing data.
#> bruceR.ID Month Day.1or2 Solar.R Wind .
#> # 5 5 Month5 odd NA 14.3 56
#> # 6 6 Month5 even NA 14.9 66
#> # 11 11 Month5 odd NA 6.9 74
#> # 27 27 Month5 odd NA 8.0 57
#> # 96 96 Month8 even NA 6.9 86
#> # 97 97 Month8 odd NA 7.4 85
#>
#> ====== ANOVA (Between-Subjects Design) ======
#>
#> Descriptives:
#> ─────────────────────────────────────
#> "Month" "Day.1or2" Mean S.D. n
#> ─────────────────────────────────────
#> Month5 odd 66.077 (7.297) 13
#> Month5 even 65.714 (6.390) 14
#> Month6 odd 78.600 (6.905) 15
#> Month6 even 79.600 (6.479) 15
#> Month7 odd 83.375 (4.064) 16
#> Month7 even 84.467 (4.642) 15
#> Month8 odd 83.667 (6.102) 15
#> Month8 even 83.846 (7.978) 13
#> Month9 odd 76.733 (9.177) 15
#> Month9 even 77.067 (7.769) 15
#> ─────────────────────────────────────
#> Total sample size: N = 153 (7 missing observations deleted)
#>
#> ANOVA Table:
#> Dependent variable(s): Temp
#> Between-subjects factor(s): Month, Day.1or2
#> Within-subjects factor(s): –
#> Covariate(s): Solar.R, Wind
#> ───────────────────────────────────────────────────────────────────────────────────
#> MS MSE df1 df2 F p η²p [90% CI of η²p] η²G
#> ───────────────────────────────────────────────────────────────────────────────────
#> Month 1059.183 35.312 4 134 29.995 <.001 *** .472 [.366, .550] .472
#> Day.1or2 6.804 35.312 1 134 0.193 .661 .001 [.000, .030] .001
#> Solar.R 612.990 35.312 1 134 17.359 <.001 *** .115 [.043, .204] .115
#> Wind 880.158 35.312 1 134 24.925 <.001 *** .157 [.074, .251] .157
#> Month * Day.1or2 7.174 35.312 4 134 0.203 .936 .006 [.000, .004] .006
#> ───────────────────────────────────────────────────────────────────────────────────
#> MSE = mean square error (the residual variance of the linear model)
#> η²p = partial eta-squared = SS / (SS + SSE) = F * df1 / (F * df1 + df2)
#> ω²p = partial omega-squared = (F - 1) * df1 / (F * df1 + df2 + 1)
#> η²G = generalized eta-squared (see Olejnik & Algina, 2003)
#> Cohen’s f² = η²p / (1 - η²p)
#>
#> Levene’s Test for Homogeneity of Variance:
#> ──────────────────────────────────────
#> Levene’s F df1 df2 p
#> ──────────────────────────────────────
#> DV: Temp 1.573 9 143 .129
#> ──────────────────────────────────────
#>
#> ------ EMMEANS (effect = "Month") ------
#>
#> Joint Tests of "Month":
#> ──────────────────────────────────────────────────────────────
#> Effect df1 df2 F p η²p [90% CI of η²p]
#> ──────────────────────────────────────────────────────────────
#> Month 4 134 29.995 <.001 *** .472 [.366, .550]
#> Day.1or2 1 134 0.193 .661 .001 [.000, .030]
#> Solar.R 1 134 17.359 <.001 *** .115 [.043, .204]
#> Wind 1 134 24.925 <.001 *** .157 [.074, .251]
#> Month * Day.1or2 4 134 0.203 .936 .006 [.000, .004]
#> ──────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "Month":
#> ──────────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ──────────────────────────────────────────────────────────────
#> Mean: "Month" 4236.730 4 1059.183 29.995 <.001 ***
#> Residuals 4731.762 134 35.312
#> ──────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "Month":
#> ────────────────────────────────────────
#> "Month" Mean [95% CI of Mean] S.E.
#> ────────────────────────────────────────
#> Month5 67.277 [64.959, 69.596] (1.172)
#> Month6 79.196 [77.048, 81.343] (1.086)
#> Month7 82.412 [80.252, 84.573] (1.092)
#> Month8 83.450 [81.201, 85.698] (1.137)
#> Month9 77.465 [75.309, 79.621] (1.090)
#> ────────────────────────────────────────
#>
#> Consecutive (Sequential) Comparisons of "Month":
#> ──────────────────────────────────────────────────────────────────────────────
#> Contrast Estimate S.E. df t p Cohen’s d [95% CI of d]
#> ──────────────────────────────────────────────────────────────────────────────
#> Month6 - Month5 11.918 (1.592) 134 7.486 <.001 *** 2.006 [ 1.327, 2.684]
#> Month7 - Month6 3.217 (1.541) 134 2.087 .155 0.541 [-0.115, 1.198]
#> Month8 - Month7 1.038 (1.573) 134 0.660 1.000 0.175 [-0.496, 0.845]
#> Month9 - Month8 -5.985 (1.571) 134 -3.808 <.001 *** -1.007 [-1.677, -0.338]
#> ──────────────────────────────────────────────────────────────────────────────
#> Pooled SD for computing Cohen’s d: 5.942
#> Results are averaged over the levels of: Day.1or2
#> P-value adjustment: Bonferroni method for 4 tests.
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
#> ------ EMMEANS (effect = "Month") ------
#>
#> Joint Tests of "Month":
#> ────────────────────────────────────────────────────────────────
#> Effect "Day.1or2" df1 df2 F p η²p [90% CI of η²p]
#> ────────────────────────────────────────────────────────────────
#> Month odd 4 134 13.278 <.001 *** .284 [.169, .371]
#> Month even 4 134 18.140 <.001 *** .351 [.236, .437]
#> Solar.R odd 1 134 17.359 <.001 *** .115 [.043, .204]
#> Solar.R even 1 134 17.359 <.001 *** .115 [.043, .204]
#> Wind odd 1 134 24.925 <.001 *** .157 [.074, .251]
#> Wind even 1 134 24.925 <.001 *** .157 [.074, .251]
#> ────────────────────────────────────────────────────────────────
#> Note. Simple effects of repeated measures with 3 or more levels
#> are different from the results obtained with SPSS MANOVA syntax.
#>
#> Univariate Tests of "Month":
#> ──────────────────────────────────────────────────────────────
#> Sum of Squares df Mean Square F p
#> ──────────────────────────────────────────────────────────────
#> odd: "Month" 1875.512 4 468.878 13.278 <.001 ***
#> even: "Month" 2562.180 4 640.545 18.140 <.001 ***
#> Residuals 4731.762 134 35.312
#> ──────────────────────────────────────────────────────────────
#> Note. Identical to the results obtained with SPSS GLM EMMEANS syntax.
#>
#> Estimated Marginal Means of "Month":
#> ───────────────────────────────────────────────────
#> "Month" "Day.1or2" Mean [95% CI of Mean] S.E.
#> ───────────────────────────────────────────────────
#> Month5 odd 67.644 [64.342, 70.946] (1.669)
#> Month6 odd 79.321 [76.275, 82.367] (1.540)
#> Month7 odd 81.991 [79.019, 84.962] (1.503)
#> Month8 odd 82.494 [79.416, 85.573] (1.557)
#> Month9 odd 77.268 [74.229, 80.307] (1.536)
#> Month5 even 66.910 [63.732, 70.089] (1.607)
#> Month6 even 79.070 [76.032, 82.109] (1.536)
#> Month7 even 82.834 [79.746, 85.922] (1.561)
#> Month8 even 84.405 [81.135, 87.676] (1.654)
#> Month9 even 77.663 [74.613, 80.713] (1.542)
#> ───────────────────────────────────────────────────
#>
#> Polynomial Contrasts of "Month":
#> ────────────────────────────────────────────────────────────
#> Contrast "Day.1or2" Estimate S.E. df t p
#> ────────────────────────────────────────────────────────────
#> linear odd 22.420 ( 5.061) 134 4.430 <.001 ***
#> quadratic odd -35.973 ( 5.946) 134 -6.050 <.001 ***
#> cubic odd 3.277 ( 4.925) 134 0.665 .507
#> quartic odd -10.405 (12.661) 134 -0.822 .413
#> linear even 26.840 ( 5.008) 134 5.359 <.001 ***
#> quadratic even -39.997 ( 5.922) 134 -6.754 <.001 ***
#> cubic even 0.082 ( 5.034) 134 0.016 .987
#> quartic even -12.327 (13.192) 134 -0.934 .352
#> ────────────────────────────────────────────────────────────
#>
#>
#> Disclaimer:
#> By default, pooled SD is Root Mean Square Error (RMSE).
#> There is much disagreement on how to compute Cohen’s d.
#> You are completely responsible for setting `sd.pooled`.
#> You might also use `effectsize::t_to_d()` to compute d.
#>
# }